Zea mays Metabolomics
Zea mays metabolomics involves the systematic study of the complete set of small molecule metabolites present in Zea mays plants. These metabolites include a diverse array of compounds such as sugars, amino acids, organic acids, lipids, secondary metabolites, and more. Metabolomics aims to comprehensively analyze and quantify these molecules to understand the biochemical reactions and pathways that govern the growth, development, responses to environmental factors, and interactions with other organisms in Zea mays.
What Zea mays Metabolomics Can Analyze?
Stress Response and Resilience: Metabolomics allows researchers to identify specific metabolites that change in response to environmental stressors such as drought, heat, cold, salinity and pathogens. By understanding these metabolic changes, key biochemical pathways involved in the stress response can be identified, contributing to the development of stress-resistant maize varieties through genetic engineering or selective breeding.
Nutritional Quality Enhancement: Metabolomics helps in assessing the nutritional composition of Zea mays and identifying metabolites associated with nutritional attributes such as vitamins, minerals, antioxidants and amino acids.
Pathway Discovery and Regulation: By analyzing metabolic pathways, you can discover how specific genes and enzymes affect metabolite production. This helps decipher the regulatory mechanisms behind maize metabolism. The insights gained can inform the development of strategies to manipulate pathways to achieve desired results, such as increased production of bioactive compounds.
Biofortification: Metabolomics contributes to biofortification efforts to add essential nutrients to crops. By identifying metabolites associated with nutrients, you can develop maize varieties enriched with specific vitamins or minerals, addressing micronutrient deficiencies in areas where maize is a staple food.
Disease and Pest Resistance: Metabolomics can identify metabolites involved in defense responses to pests and pathogens, helping to breed maize varieties that are more resistant to pests and diseases, thereby reducing the need for chemical pesticides.
Phenotypic Analysis: Metabolomics provides a phenotypic snapshot of Zea mays plants under different conditions. It complements genotypic and transcriptomic data by offering insights into the actual biochemical products of genes. This information aids in understanding genotype-phenotype relationships and can uncover hidden metabolic variation among different Zea mays varieties.
Environmental Adaptation: Metabolomics can help reveal how Zea mays adapts to different environments. You can identify metabolites that are associated with specific environmental conditions, thereby revealing the ability of maize to grow in different climates and soils.
Marker Discovery for Breeding: Metabolomics can identify metabolite markers associated with specific traits, marker-assisted breeding programs, and more effective screening of Zea mays plants for desired traits.
Post-Harvest Quality: Metabolomics can help understand post-harvest quality changes in Zea mays, identifying metabolites that affect shelf life, taste, texture, and overall consumer satisfaction to help develop storage and preservation strategies.
Climate Change Adaptation: Metabolomics provides insights into how maize is responding to climate change, including changes in temperature, carbon dioxide levels, and precipitation patterns, informing strategies for developing climate-resilient maize varieties.
Zea mays Metabolomics Analysis Techniques
Liquid Chromatography-Mass Spectrometry (LC-MS): LC-MS combines liquid chromatography separation with mass spectrometric detection. It facilitates separation and precise quantification of diverse metabolites in complex mixtures.
Instrument Model: Agilent 1290 Infinity LC coupled with Agilent 6550 iFunnel Q-TOF Mass Spectrometer.
Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS is used for analyzing volatile and evaporative metabolites. Gas chromatographic separation is followed by mass spectrometric detection and quantification.
Instrument Model: Agilent 7890A GC coupled with Agilent 5977A MSD.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS): LC-MS/MS combines liquid chromatography with tandem mass spectrometry. It provides enhanced accuracy for metabolite quantification and structural identification.
Instrument Model: Thermo Scientific Q Exactive Plus.
High-Resolution Mass Spectrometry (HRMS): HRMS offers precise mass measurement and accurate quantification of metabolites.
Instrument Model: Thermo Scientific Orbitrap series.
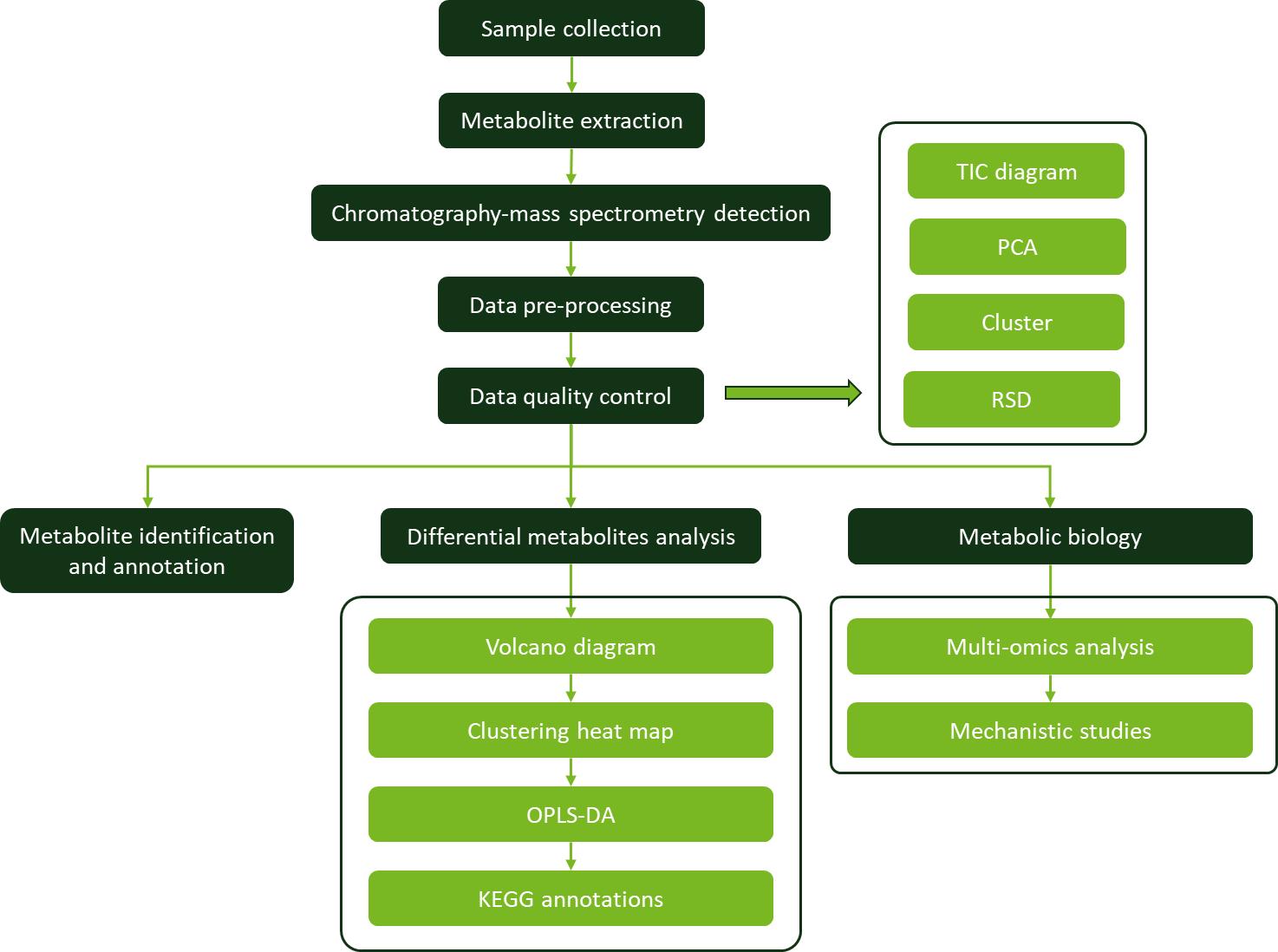 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Sample Requirements for Zea mays Metabolomics
| Sample Type |
Sample Requirements and Recommended Sample Sizes |
| Leaves |
- Collect leaves from multiple plants at the same developmental stage. |
| - Each sample should contain enough leaves for reliable metabolite analysis. |
| - Recommended sample size: 50-100 grams (fresh weight). |
| Stems |
- Collect stems at different heights from the same plant to cover the entire stem. |
| - Consider stems at various developmental stages and under different conditions. |
| - Recommended sample size: 20-50 grams (fresh weight). |
| Seeds (Kernels) |
- Collect kernels at different developmental stages, from early to mature. |
| - Account for variations in seed size, morphology, and chemical composition. |
| - Recommended sample size: 20-30 grams (fresh weight). |
| Roots |
- Collect root samples from deeper soil layers to represent different growth depths. |
| - Consider variations in root metabolism at different growth stages and environmental conditions. |
| - Recommended sample size: 10-30 grams (fresh weight). |
Case 1. Exploring Maize Metabolomic Responses to Drought Stress: Insights from Extraction Techniques and Tissue Analysis
Background:
Drought stress poses a significant threat to global food security, particularly impacting crops like maize (Zea mays L.). Metabolomics, a comprehensive analysis of a biological sample's chemical composition, has been crucial in understanding plant responses to stress. In this context, the study focuses on the influence of drought stress on maize metabolites, considering their role in adaptation and signaling under changing environmental conditions.
Samples:
The study employed the B73 variety of maize plants, known for drought susceptibility. Two main types of samples were investigated: mature corn cobs and kernels. The experiment included both an "extraction condition" phase and a "drought condition" phase. The former compared various extraction techniques, including liquid nitrogen-based extraction and lyophilization, to stabilize the plant metabolome for subsequent analysis. The latter involved growing maize plants under different watering conditions and harvesting cobs at maturity for metabolomic analysis.
Methods:
Sample Preparation:
Two main phases were conducted: the "extraction condition" experiment and the "drought condition" experiment. The "extraction condition" phase involved preparing maize cob tissue samples using three extraction methods: liquid nitrogen extraction (LNE), lyophilized cold extraction (LC), and lyophilized room temperature extraction (LRT). The "drought condition" phase involved growing maize plants under controlled conditions, subjecting them to varying drought stress levels, and harvesting cobs at maturity. Samples from both kernel and inner cob tissues were collected for analysis.
Metabolite Extraction and LC–MS Analysis:
Metabolite extraction was performed using a methanol:water (70:30, v/v) solvent mixture. The extraction process included shaking the samples for 30 minutes in the dark, followed by centrifugation. The resulting supernatant was dried using a vacuum concentrator, and the extracted metabolites were reconstituted in methanol:water. High-performance liquid chromatography (HPLC) was employed to separate the metabolites based on polarity using an Atlantis® T3 column. The HPLC was coupled to an electrospray Fourier-transform ion cyclotron resonance mass spectrometer (FT-ICR–MS) for mass analysis.
Data Collection and Analysis:
For the "extraction condition" experiment, data processing was done using Progenesis QI software. Raw data were imported in centroid mode, and alignment was performed based on reference runs. Peak picking parameters were adjusted to reduce data volume. Features related to charge-bearing species were considered. Statistical analyses involved one-way analysis of variance (ANOVA) to determine differences between groups. The "drought condition" experiment followed similar data processing steps, and multivariate techniques such as principal components analysis (PCA) were applied to analyze the data.
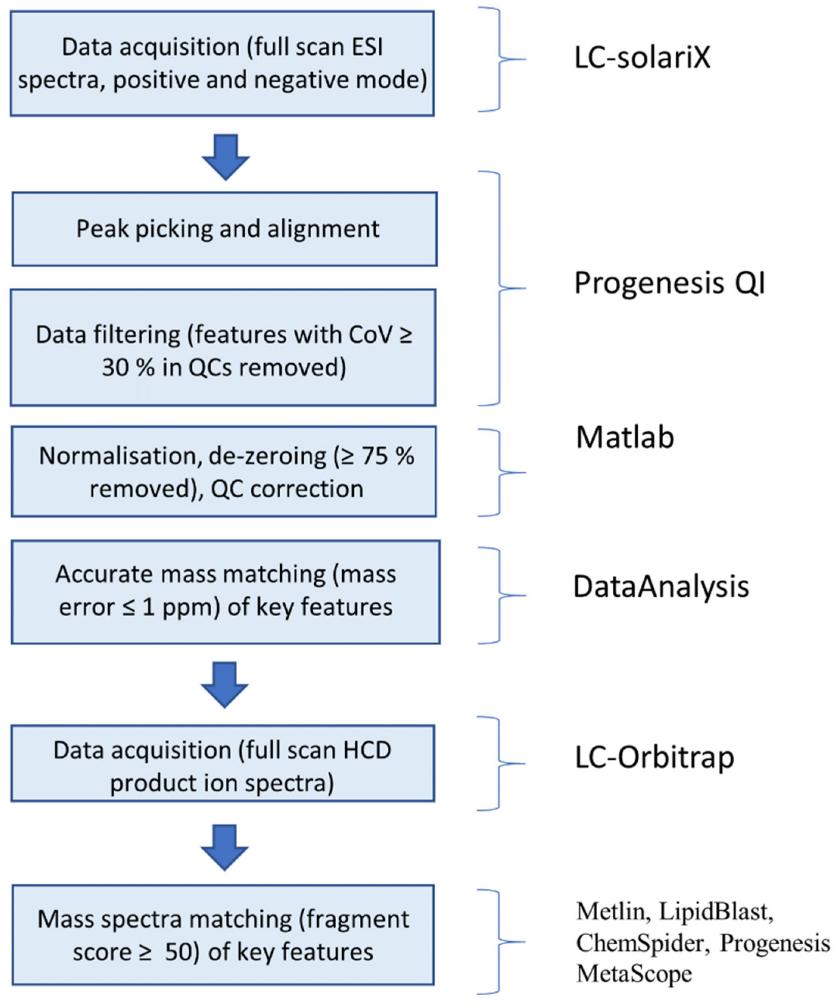 Data analysis workflow for the identification of key features in mass spectrometric data.
Data analysis workflow for the identification of key features in mass spectrometric data.
Results
The study revealed that lyophilization is a practical and effective alternative to liquid nitrogen-based extraction, facilitating the stabilization of the extractable plant metabolome in maize samples. This is significant for both resource efficiency and safety considerations in sample processing. Furthermore, the analysis of drought-stressed maize samples uncovered a strong metabolic response, with significant changes observed in metabolite profiles between well-watered and drought-stressed conditions. The divergence in metabolomic profiles was most pronounced when drought stress was introduced at the three-leaf stage, reflecting the plants' adaptation to changing conditions. Inner cob tissue also emerged as a valuable source of information in cases where kernel material might be limited, highlighting the utility of non-targeted metabolomics in uncovering the mechanisms of plant stress responses.
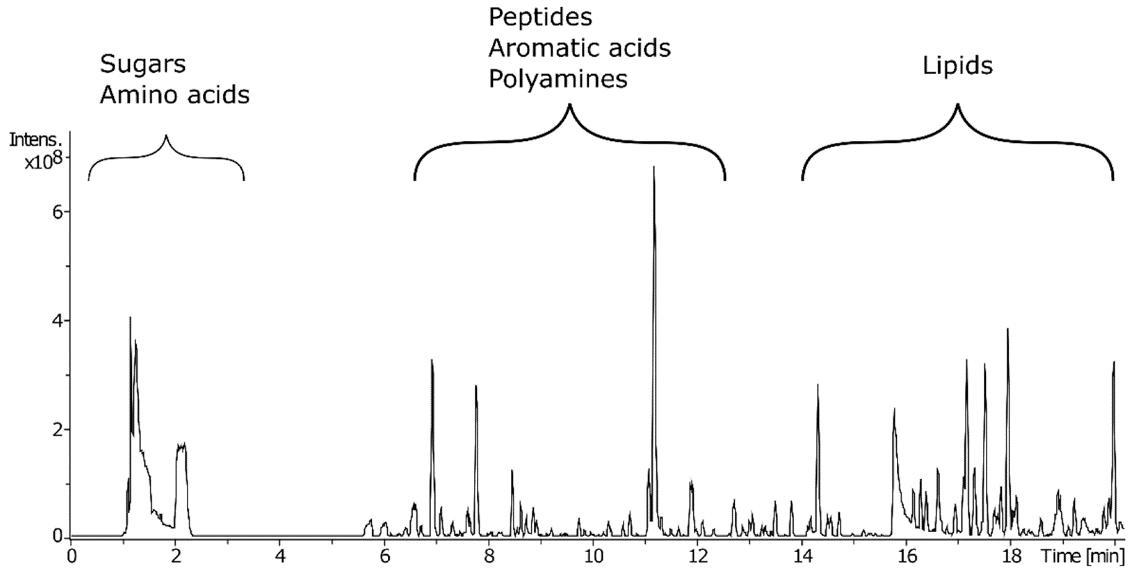 Characteristic base peak ion chromatogram for an LNE sample obtained using a T3 column coupled to high-resolution mass spectrometry (LC–HRMS) to allow the retention and separation of metabolites with a broad range of polarities.
Characteristic base peak ion chromatogram for an LNE sample obtained using a T3 column coupled to high-resolution mass spectrometry (LC–HRMS) to allow the retention and separation of metabolites with a broad range of polarities.
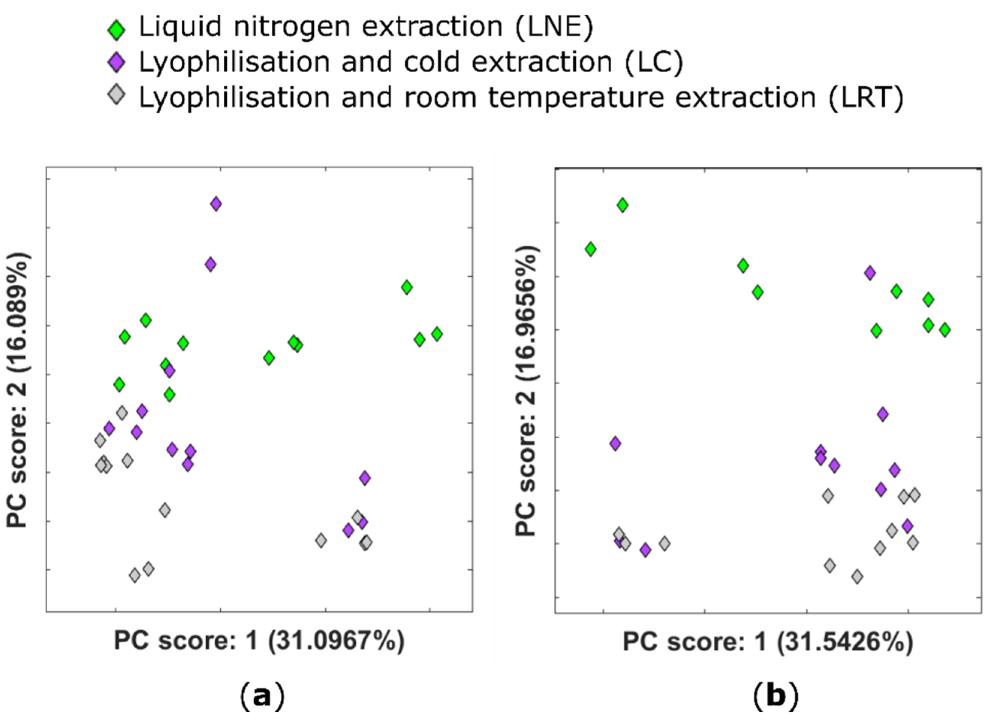 PCA plots showing the scores for the first two principal components obtained using untargeted metabolomic analysis of different extraction conditions for Zea mays
PCA plots showing the scores for the first two principal components obtained using untargeted metabolomic analysis of different extraction conditions for Zea mays
Reference
- Gaffney, Isabella, et al. "Metabolomic approaches to studying the response to drought stress in corn (Zea mays) cobs." Metabolites 11.7 (2021): 438.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service Data analysis workflow for the identification of key features in mass spectrometric data.
Data analysis workflow for the identification of key features in mass spectrometric data. Characteristic base peak ion chromatogram for an LNE sample obtained using a T3 column coupled to high-resolution mass spectrometry (LC–HRMS) to allow the retention and separation of metabolites with a broad range of polarities.
Characteristic base peak ion chromatogram for an LNE sample obtained using a T3 column coupled to high-resolution mass spectrometry (LC–HRMS) to allow the retention and separation of metabolites with a broad range of polarities. PCA plots showing the scores for the first two principal components obtained using untargeted metabolomic analysis of different extraction conditions for Zea mays
PCA plots showing the scores for the first two principal components obtained using untargeted metabolomic analysis of different extraction conditions for Zea mays

