Arachis hypogaea Metabolomics
Arachis hypogaea, commonly referred to as peanuts, holds global significance as a vital crop due to its economic and nutritional contributions. Metabolomics, a potent analytical method, provides an encompassing understanding of the intricate metabolic mechanisms operating within peanuts, thereby yielding valuable insights into growth patterns, responses to stressors, and nutritional composition.
Metabolomics delves into the exploration of small molecule metabolites inherent to biological systems. By employing this approach, researchers can concurrently scrutinize an extensive array of metabolites, encompassing carbohydrates, amino acids, lipids, and secondary metabolites. This capability facilitates a comprehensive grasp of peanut metabolism, enhancing our comprehension of its intricate biochemical pathways.
Our Metabolomic Service for Arachis hypogaea Which Research
Growth and Development: Metabolomics helps understand the metabolic changes in peanuts across different growth stages, revealing the involved pathways and key molecules during growth and development.
Stress Response: In the face of adversities such as drought, high temperatures, and pests, metabolomics analyzes changes in metabolic pathways, uncovering stress response mechanisms and aiding the development of stress-tolerant varieties.
Quality and Nutrition: Metabolomics evaluates peanut quality and nutritional components, including proteins, amino acids, lipids, and vitamins. This aids in refining cultivation strategies to enhance yield and quality.
Gene Function Research: Metabolomics helps study gene function in peanuts, exploring the role of specific genes in metabolic regulation through the analysis of metabolite changes in mutants or transgenic plants.
Genetic Diversity: Metabolomics can compare metabolic differences among different peanut varieties, revealing the impact of genetic diversity on metabolic traits and informing the breeding of high-quality varieties.
Ecological Interactions: Metabolomics explores interactions between peanuts and microbes, soil, and other organisms, revealing symbiotic relationships' effects on peanut metabolism.
Arachis hypogaea Metabolomics Analysis Project at Creative Proteomics
Metabolite Profiling: We conduct global profiling of metabolites, including carbohydrates, amino acids, lipids, and secondary metabolites, providing a broad view of peanut metabolism.
Metabolite Identification: Our advanced techniques identify and annotate metabolites, enabling precise characterization of the detected compounds in peanuts.
Pathway Analysis: We perform pathway enrichment and mapping to understand how metabolites are interconnected within different metabolic pathways in peanuts.
Comparative Metabolomics: We offer comparative analysis of different peanut varieties or growth conditions, highlighting metabolic differences and genetic diversity effects.
Biomarker Discovery: Metabolomics aids in identifying potential biomarkers for quality, stress tolerance, and other desirable traits in peanuts.
Metabolic Flux Analysis: Our experts analyze the flow of metabolites within different metabolic pathways, providing insights into metabolic fluxes and regulatory mechanisms.
Multivariate Statistical Analysis: We employ advanced statistical methods to analyze complex metabolomics data, identifying significant patterns and correlations.
Arachis hypogaea Metabolomics Analysis Techniques
Liquid Chromatography-Mass Spectrometry (LC-MS):
LC-MS is a versatile approach that combines liquid chromatography for separation and mass spectrometry for detection. It enables the identification and quantification of a wide range of polar and nonpolar metabolites. The high sensitivity and selectivity of LC-MS make it suitable for analyzing complex mixtures of metabolites in peanut samples. By utilizing instrument models such as the Agilent 6550 Q-TOF, Thermo Scientific Q Exactive HF-X, and Waters Xevo G2-XS QTOF, researchers can gain insights into the diverse metabolites present in peanuts and how they respond to various conditions.
Gas Chromatography-Mass Spectrometry (GC-MS):
GC-MS is particularly well-suited for analyzing volatile and thermally stable compounds. In this technique, metabolites are first derivatized to enhance their volatility and then subjected to gas chromatographic separation followed by mass spectrometric detection. Instrument models such as the Agilent 7890B GC coupled with Agilent 5977A MSD, Thermo Scientific ISQ GC-MS, and Shimadzu GCMS-QP2020 NX are commonly employed for GC-MS analysis. GC-MS allows researchers to explore the composition of fatty acids, amino acids, sugars, and other metabolites critical for peanut metabolism.
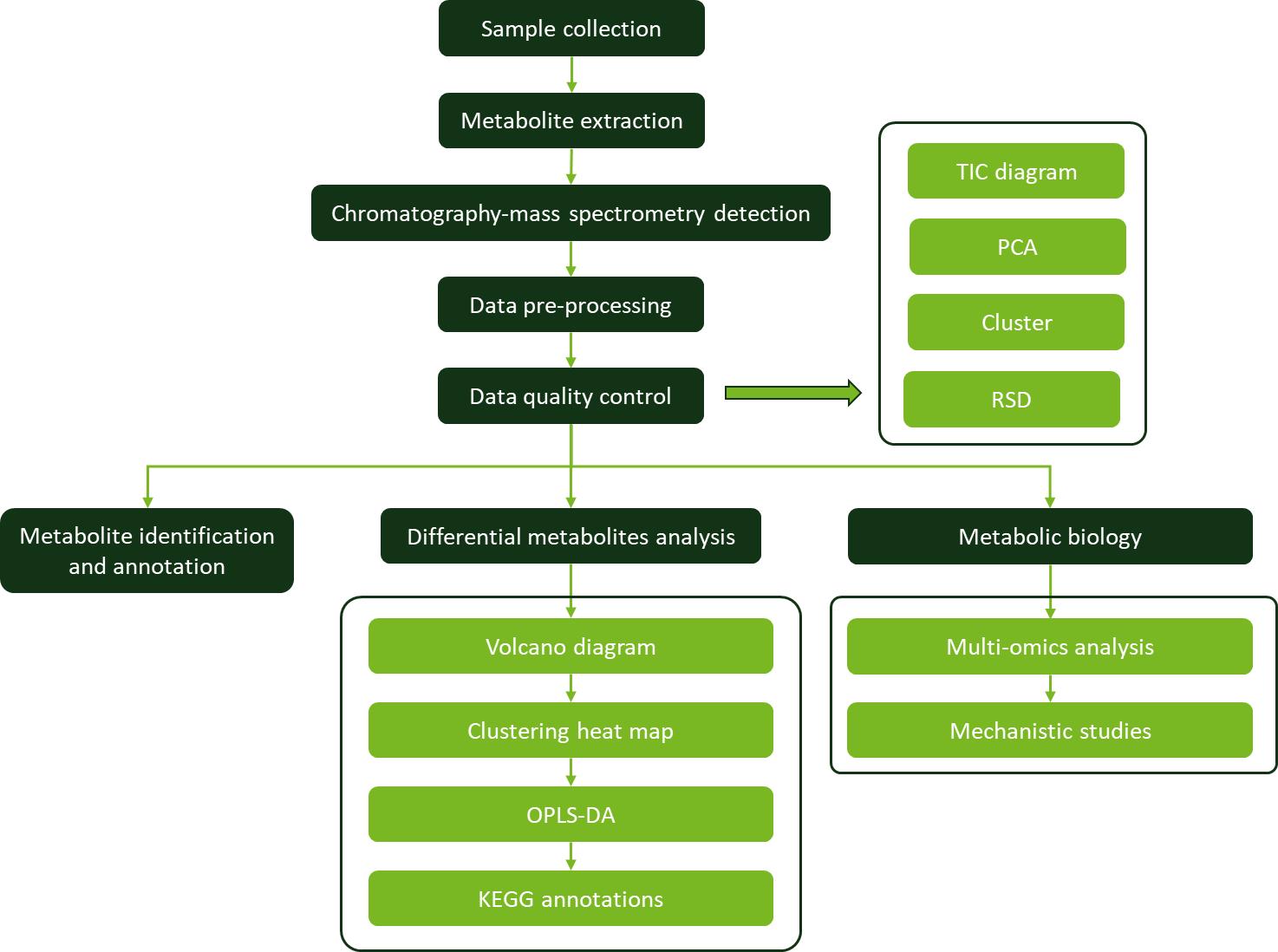 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Sample Requirements for Arachis hypogaea Metabolomics
| Sample Type |
Sample Amount |
Biological Replicates |
| Plant Tissues |
100 mg per tissue |
3-5 replicates |
| Roots |
50 mg per sample |
3 replicates |
| Flowers |
100 mg per sample |
3 replicates |
| Fruits |
100 mg per sample |
3 replicates |
| Nodules (Root Bumps) |
50 mg per sample |
3 replicates |
| Different Growth Stages |
100 mg per sample |
3 replicates |
| Varied Environmental Conditions |
100 mg per sample |
3 replicates |
| Stress-treated Samples |
100 mg per sample |
3 replicates |
| Wild Type vs. Transgenic |
100 mg per sample |
3 replicates |
Case 1. Integrated Transcriptomic and Metabolomic Analysis Reveals Molecular Mechanisms Underlying Color Variation in Variegated Peanut Testa
Background:
Peanut testa color variation is primarily determined by anthocyanin pigments. Investigating the genetic and molecular basis of pigment synthesis is crucial for enhancing peanut breeding and resource preservation. This study seeks to unravel the intricate mechanisms driving color differences in the variegated peanut testa through an integrated approach involving transcriptomics and metabolomics.
Samples:
The study employed variegated peanut testa, sourced from accession VG-01. This material exhibits distinct pigmented (red) and non-pigmented (white) areas. Seeds were procured from the Peanut Breeding Laboratory at Hebei Agricultural University, China. Samples were meticulously collected at various days post flowering and needling (DAF) to capture the dynamic developmental stages of the testa.
Methods:
Characterization and Chromatic Aberration: The stereomicroscope was used to observe color changes in the testa. Chromatic aberration values (L, a, b) were measured using a colorimeter, offering quantifiable insights into color variations at different DAF stages.
Flavonoids Extraction: Testa samples were pulverized and subjected to overnight extraction in 70% aqueous methanol. The resulting extracts were absorbed, filtered, and analyzed using liquid chromatography-tandem mass spectrometry (LC–MS).
Species and Quantitative Analysis of Flavonoids: LC–MS/MS was employed for qualitative and quantitative analysis of metabolites in different testa areas. The technique involved dissolving and diluting testa samples before mass spectrometry analysis with ultra-high-performance liquid chromatography.
Transcriptome Sequencing and DEGs Analysis: Illumina sequencing was conducted on the samples, with comparison against diploid wild and Tifrunner genomes. Differential gene expression analysis employed DEseq software, with specific criteria to identify differentially expressed genes (DEGs).
GO and KEGG Analysis of DEGs: DEGs were subjected to gene ontology (GO) and KEGG pathway enrichment analysis. This step aimed to understand the biological functions and metabolic pathways associated with DEGs.
Joint Analysis of Transcriptome and Metabolome: Integration of transcriptome and metabolome data involved Pearson correlation analysis to identify relationships between differential genes and metabolites.
Verification by qRT-PCR: A subset of 20 DEGs was selected for verification using quantitative real-time PCR (qRT-PCR). RNA extraction, cDNA synthesis, and primer design were performed, followed by gene expression analysis using the ΔΔCt method.
Results
The comprehensive analysis highlighted several pivotal findings. The investigation identified key genes involved in pigment synthesis, including CHS and DFR, as well as regulatory genes like bHLH and MYB. Joint analysis of transcriptome and metabolome data provided strong evidence of the tight integration of flavonoid biosynthesis pathways. Notably, the accumulation of delphinidin and cyanidin emerged as the primary factors driving color distinctions between pigmented and non-pigmented regions. This knowledge of the molecular regulation of pigment synthesis holds significant implications for the advancement of peanut breeding strategies and the preservation of valuable agricultural resources.
Reference
- Hu, Mengdie, et al. "Transcriptomic and metabolomic joint analysis reveals distinct flavonoid biosynthesis regulation for variegated testa color development in peanut (Arachis hypogaea L.)." Scientific Reports 11.1 (2021): 10721.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service

