GC-MS metabolomics is applicable for qualitative and quantitative analysis of unknown components in complex mixtures. It allows for the determination of molecular structures and accurate measurement of molecular weights, making it suitable for trace analysis of metabolites in plant samples. For volatile components, direct qualitative and quantitative analysis is feasible, while for non-volatile and thermally unstable substances, appropriate derivatization can be performed prior to analysis. With advantages such as high sensitivity, robust qualitative capabilities, and reliability, GC-MS enables the simultaneous separation and identification of target components. The application of electron ionization in GC-MS results in the generation of extensive and highly reproducible fragmentation patterns. Even when mass spectral data is not available in databases, these fragmentation patterns can be utilized to obtain additional information about the qualitative aspects or types of metabolites or compounds.
Types of GC-MS Instruments
- Single Quadrupole GC-MS: Suitable for routine analyses, offering robust performance.
- Triple Quadrupole GC-MS: Ideal for targeted compound quantification with enhanced sensitivity.
- Time-of-Flight (TOF) GC-MS: Provides high-resolution mass spectrometry for accurate mass determination.
Creative Proteomics is a leading provider of comprehensive GC-MS plant metabolomics services. With a commitment to precision and innovation, our GC-MS plant metabolomics services facilitate both qualitative and quantitative analyses of unknown components within complex mixtures in plant samples. Leveraging the sensitivity, robust qualitative capabilities, and reliability of GC-MS technology, Creative Proteomics delivers accurate insights into the molecular structures and weights of metabolites present in plants. Our services cover a broad spectrum, from trace analysis of volatile components to the examination of non-volatile and thermally unstable substances through suitable derivatization methods. With a focus on high-standardized electron ionization applications, our GC-MS services generate extensive and reproducible fragmentation patterns, providing valuable information even when mass spectral data is not available in databases.
Advantages of GC-MS Technology
Wide Analytical Range: GC-MS excels in detecting diverse compounds, addressing challenges with thermally unstable substances. This capability ensures comprehensive compound identification.
Strong Separation Capability: In situations lacking complete chromatogram separation, MS enables effective component differentiation. This facilitates precise qualitative and quantitative analyses.
Reliable Qualitative Analysis: Simultaneous molecular weight and structural information by GC-MS ensure reliable qualitative analysis, revealing compound identity and structure.
Low Detection Limits: High sensitivity and Selected Ion Monitoring (SIM) in GC-MS significantly lower detection limits, identifying trace amounts for enhanced accuracy.
Rapid Analysis: Efficient gas chromatography with narrow-bore columns reduces analysis time in GC-MS, allowing high-throughput studies without compromising data quality.
High Automation Level: GC-MS's automation streamlines processes, enhancing efficiency and minimizing human error for reproducible and reliable results.
Targeted GC-MS Metabolomics Analysis:
Quantitative Analysis:
- Specific Metabolite Measurement: Targeted plant metabolomics services focuses on the quantification of specific known metabolites. GC-MS allows for the precise measurement of individual metabolites, providing accurate concentration information.
- Internal Standards: The use of internal standards facilitates quantitative analysis by correcting for variations in sample preparation and instrument response. This ensures the accuracy and reproducibility of the results.
Pathway Analysis:
- Metabolic Pathway Mapping: Targeted approaches often involve studying specific metabolic pathways. GC-MS aids in identifying and quantifying metabolites involved in pathways such as glycolysis, tricarboxylic acid (TCA) cycle, and amino acid metabolism.
- Enzyme Activity Assessment: By measuring specific metabolites, researchers can infer the activity of enzymes involved in particular pathways, providing insights into metabolic regulation.
Untargeted Metabolomics Analysis:
Comprehensive Profiling:
- Identification of Unknown Metabolites: Untargeted plant metabolomics aims to capture the entire metabolome without prior knowledge of specific metabolites. GC-MS, coupled with sophisticated data analysis techniques, allows for the identification of unknown metabolites.
- Global Metabolic Changes: Researchers can observe global changes in the plant metabolome under different conditions, uncovering novel biomarkers or pathways associated with specific physiological responses.
Pattern Recognition:
- Multivariate Data Analysis: Untargeted strategies involve the application of multivariate statistical methods to analyze complex datasets. Principal Component Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA) help identify patterns and groupings in metabolite data.
- Cluster Analysis: Hierarchical clustering or k-means clustering can be employed to group metabolites with similar expression patterns, aiding in the identification of co-regulated metabolic pathways.
Integration of Targeted and Untargeted Strategies:
- Holistic Understanding: Combining both targeted and non-targeted approaches provides a holistic view of plant metabolism. Targeted analysis offers specific quantitative data, while non-targeted analysis captures the complexity of the entire metabolome.
- Validation and Confirmation: Non-targeted findings can be validated using targeted approaches, ensuring the reliability of observed metabolic changes.
Data Interpretation and Reporting Service
Data Interpretation
- Peak Identification and Quantification: Advanced software is utilized for peak identification, determining the presence and concentration of each compound. Quantitative data offers precise information on the relative or absolute content of each identified compound.
- Peak Calibration and Library Matching: Peaks are calibrated, and each compound's peak is matched against a previously established mass spectral library. Library matching ensures accurate compound identification, providing reliable analytical results.
Report Generation
- Report Types: We offer customized report types to meet the specific needs of your research or project. Common report types include quantitative analysis reports, mass spectra profile reports, and comprehensive analysis reports.
- Report Formats: Reports are presented in a clear, reader-friendly format to ensure easy understanding and utilization of the information. We support multiple formats, including PDF, Word, and HTML, to accommodate your preferences and sharing requirements.
- Report Content: Reports include detailed peak tables listing each detected compound and its relative or absolute content. Information such as compound names, peak areas, retention times, etc., is presented clearly for a comprehensive understanding.
- Result Interpretation: Result interpretation is provided to help you understand the compound characteristics reflected in the GC-MS data and their distribution in the samples. Interpretation covers potential biological implications, offering deeper insights for your research.
- Graphics and Spectra: Reports include intuitive graphs and mass spectra profiles to support the visual understanding of the data. Graphics highlight key trends, differences between samples, and important compound features.
Case. Metabolomic Profiling of Volatile Organic Compounds Secreted by Plant Growth-Promoting Rhizobacteria
Background
Plant Growth-Promoting Rhizobacteria (PGPR) play a crucial role in enhancing plant growth and providing protection against pathogens. Understanding the specific mechanisms of PGPR, particularly the diverse volatile organic compounds (VOCs) they secrete, is essential for harnessing their potential in sustainable agriculture. This study focuses on the metabolomic profiling of VOCs to uncover strain-specific features influencing plant-microbe interactions.
Samples
Four strains of PGPR were investigated: Ps. fluorescens (N04), Ps. koreensis (N19), Pa. alvei (T19), and L. sphaericus (T22). The study aimed to compare their volatile profiles and identify unique characteristics contributing to their plant growth-promoting and pathogen-inhibiting capabilities.
Technological Methods
The study employed high-resolution GC–HR–time-of-flight–MS (Gas Chromatography-High Resolution-Time of Flight-Mass Spectrometry) for metabolomic analysis of the VOCs. This technology allows for concurrent detection of multiple analytes with high sensitivity. Data pre-processing and chemometric techniques were utilized to extract underlying features contributing to observed differences in volatile profiles. Principal Component Analysis (PCA), Hierarchical Cluster Analysis (HCA), and Orthogonal Projection to Latent Discriminant Analysis (OPLS-DA) were employed for data exploration, clustering, and identification of significant features.
Results
The results revealed complex and strain-specific VOC profiles among the investigated PGPR strains. PCA showed distinct groupings, with Ps. koreensis (N19) and Ps. fluorescens (N04) clustering closely, while L. sphaericus (T22) exhibited a unique profile. HCA further divided strains into clusters, emphasizing differences in volatile profiles. OPLS-DA models were used to extract features responsible for dissimilarities, and permutation tests validated model predictability.
In total, 121 VOCs were identified, belonging to various classes such as aldehydes, alcohols, ketones, acids, and terpenoids. The comparison of Pseudomonas strains revealed shared and unique VOCs, including plant hormone derivatives like methyl salicylate. Additionally, the study highlighted Pa. alvei (T22) as having the highest number of unique VOCs, potentially contributing to its efficacy in inducing/enhancing resistance against phytopathogens.
This research underscores the specificity of VOC profiles among PGPR strains and emphasizes the potential of metabolomic approaches for deeper insights into their plant-protective mechanisms.
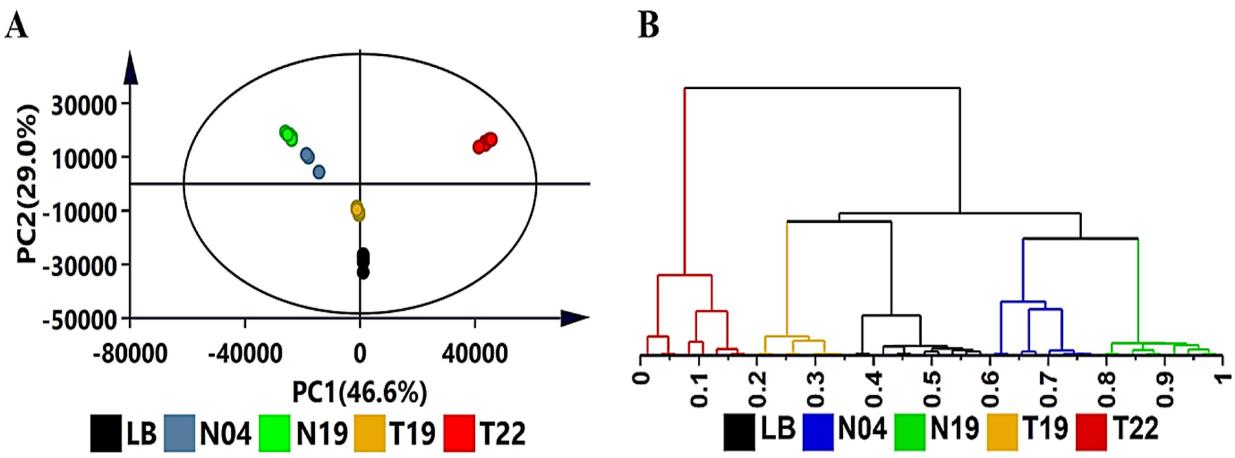 Exploratory data analysis of volatile organic compounds produced by the four PGPR strains (N19, N04, T19, T22) with unsupervised chemometric methods.
Exploratory data analysis of volatile organic compounds produced by the four PGPR strains (N19, N04, T19, T22) with unsupervised chemometric methods.
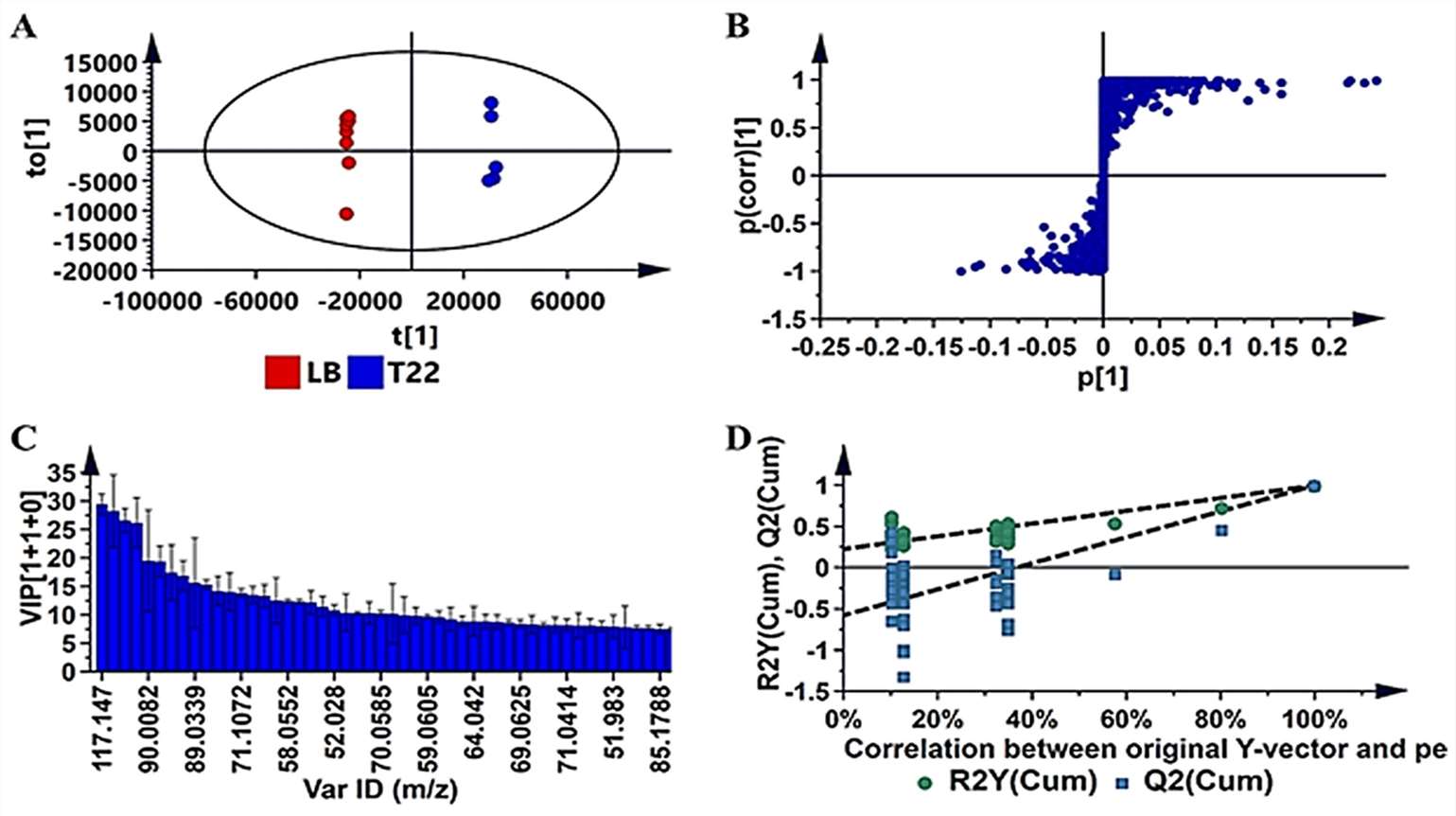 OPLS-DA modeling and variable/feature selection of Lysinibacillus sphaericus (T22) data acquired on GC–TOF–MS.
OPLS-DA modeling and variable/feature selection of Lysinibacillus sphaericus (T22) data acquired on GC–TOF–MS.
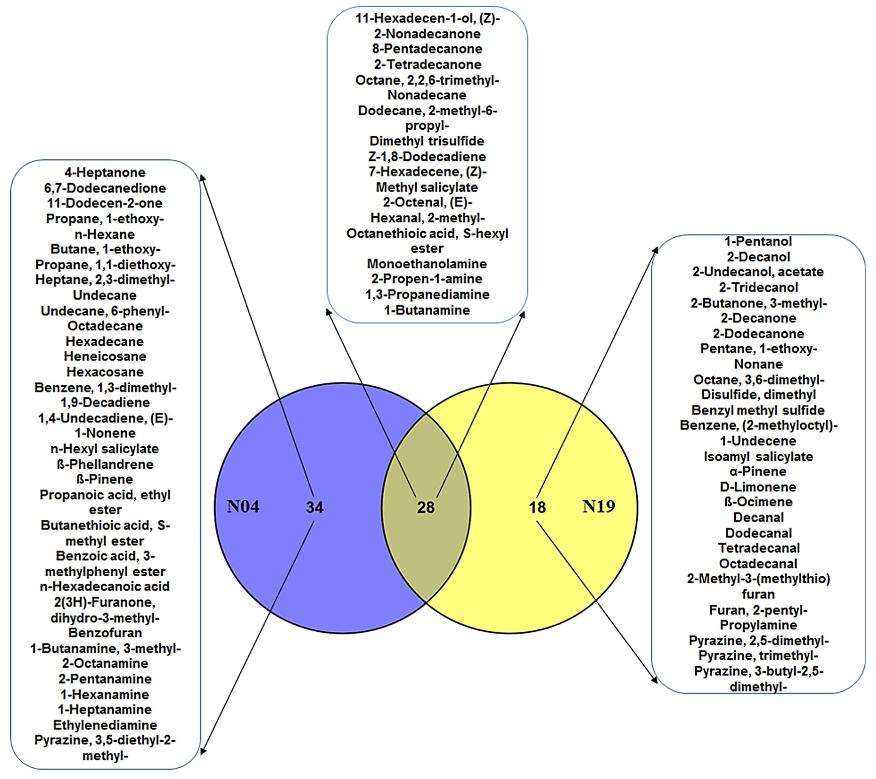 Venn diagram comparing the volatile organic compounds of the two Pseudomonas strains.
Venn diagram comparing the volatile organic compounds of the two Pseudomonas strains.
Reference
- Mhlongo, Msizi I., Lizelle A. Piater, and Ian A. Dubery. "Profiling of volatile organic compounds from four Plant Growth-Promoting Rhizobacteria by SPME–GC–MS: A metabolomics study." Metabolites 12.8 (2022): 763.


 Exploratory data analysis of volatile organic compounds produced by the four PGPR strains (N19, N04, T19, T22) with unsupervised chemometric methods.
Exploratory data analysis of volatile organic compounds produced by the four PGPR strains (N19, N04, T19, T22) with unsupervised chemometric methods. OPLS-DA modeling and variable/feature selection of Lysinibacillus sphaericus (T22) data acquired on GC–TOF–MS.
OPLS-DA modeling and variable/feature selection of Lysinibacillus sphaericus (T22) data acquired on GC–TOF–MS. Venn diagram comparing the volatile organic compounds of the two Pseudomonas strains.
Venn diagram comparing the volatile organic compounds of the two Pseudomonas strains.

