What is Disaccharide Metabolism?
Disaccharides, formed through the linkage of two monosaccharide units by glycosidic bonds, constitute essential components of plant physiology. They serve as pivotal energy reservoirs, structural constituents, and signaling mediators. Prominent disaccharides found in plants include sucrose, lactose, and maltose, with sucrose being the most abundant. Functioning as the primary conduit for assimilated carbon, sucrose facilitates nutrient translocation from photosynthetic source tissues, such as leaves, to non-photosynthetic sink tissues, encompassing roots, fruits, and seeds.
The metabolic pathways governing disaccharides are integral to plant biology, playing critical roles in energy mobilization, carbon partitioning, stress adaptation, and the orchestration of growth and development across diverse plant species. A comprehensive exploration of the intricate mechanisms underlying disaccharide metabolism not only advances our comprehension of fundamental plant physiology but also bears profound implications for enhancing agricultural productivity and fostering sustainable agricultural paradigms.
Disaccharide Metabolism Analysis Service by Creative Proteomics
Qualitative Characterization of Disaccharides and Disaccharide Metabolites: At Creative Proteomics, disaccharides and their metabolites undergo qualitative characterization through advanced methods. Our techniques include Ultra-performance liquid chromatography/multiple reaction monitoring mass spectrometry (UPLC-MRM/MS) and gas chromatography-mass spectrometry (GC-MS). Additionally, NMR spectroscopy serves as a pivotal tool for discerning the types of sugar compounds. Varied sugar compounds yield unique NMR spectra, thereby simplifying qualitative identification through spectral analysis against standard references.
Quantitative Characterization of Disaccharides and Disaccharide Metabolites: For the quantitative characterization of disaccharides and their metabolites, we employ specialized enzymes to catalyze the hydrolysis of disaccharides into individual monosaccharides, enabling precise quantification. Simultaneously, LC-MS/MS or GC-MS techniques are utilized to quantify the resultant monosaccharides. Concentration determination of carbohydrates within the sample relies on calibration curves derived from standard materials of known concentrations.
Customized Disaccharide Metabolic Profiling Analysis: At Creative Proteomics, we offer tailored solutions to explore the dynamic changes in plant disaccharide metabolites across diverse growth and developmental stages as well as varying environmental conditions.
List of Disaccharides and Disaccharide Metabolites (including but not limited to)
| Disaccharides |
Abbreviation |
| Sucrose |
Suc |
| Lactose |
Lac |
| Maltose |
Mal |
| Trehalose |
Tre |
| Cellobiose |
Cel |
| Sophorose |
Sph |
| Xylobiose |
Xyl |
| Lactitol |
Lct |
Techniques and Instrumentation for Disaccharide Metabolism Analysis
Liquid Chromatography-Mass Spectrometry (LC-MS): Utilizing advanced instruments such as the Waters ACQUITY UPLC and AB Sciex TripleQuad 5500, we conduct precise qualitative and quantitative analyses of various disaccharides and their metabolites. LC-MS technology provides comprehensive insights into metabolic processes.
Gas Chromatography-Mass Spectrometry (GC-MS): Additionally, we utilize the Agilent 7890A-5975C instrument for analyzing disaccharides and their metabolites. GC-MS technology offers exceptional selectivity and sensitivity, enabling effective differentiation of isomers in samples and ensuring the generation of accurate data crucial for advancing research in disaccharide metabolism.
Nuclear Magnetic Resonance Spectrometry (NMR): Our capabilities extend to NMR spectroscopy, with access to the Varian VNMRS 600 MHz instrument. NMR spectroscopy provides valuable insights into structural elucidation and qualitative analysis of disaccharide and their metabolites.
Why Choose Us?
- Pre-Derivatization Processing (Optional): Our service provides the option of pre-derivatization processing, ensuring optimal sample preparation tailored to your specific requirements. This step enhances the accuracy and sensitivity of our analysis, facilitating precise identification and quantification of disaccharide components.
- Accurate Quantification and Identification: Creative Proteomics utilizes standard reference materials for both qualitative and quantitative analysis of disaccharides. By employing validated analytical methods and state-of-the-art instrumentation, we ensure accurate determination of disaccharide concentrations in your samples.
- Customized Analysis Tailored to Specific Research Needs: At Creative Proteomics, we understand that each research project is unique, and thus we offer customized disaccharide metabolism analysis tailored to your specific research needs. Whether you require specialized sample preparation techniques, targeted analysis of specific disaccharides, or customized reporting formats, our team of experts will work closely with you to design an analysis strategy that meets your exact requirements.
- Comprehensive Data Analysis: Our service includes comprehensive data analysis to provide valuable insights into your research. This involves employing advanced techniques for raw data pre-processing, sophisticated multivariate analysis methods. In addition, through our commercial metabolite libraries and self-built chemical libraries, effective disaccharide metabolites distribution difference/enrichment analysis can be achieved.
Applications of Disaccharide Metabolism Analysis
- Quantitative Detection of Plant Disaccharides: We provide valuable information on the composition and dynamics of disaccharides in plants, aiding in quantitative detection.
- Regulation Pathways of Plant Sugar Metabolism: Our analysis service offers insights into the intricate regulatory mechanisms governing sugar metabolism pathways in plants.
- Understanding the Interactions between Sugars and Other Plant Hormones: By elucidating these interactions, researchers gain a deeper understanding of how sugars influence plant growth, development, and response to environmental stimuli.
- Regulation of Plant Growth and Development: Metabolite differential analysis can help researchers discover important enzymes and signaling molecules in the pathway, and provide a reference for in-depth understanding of the transport, decomposition, and synthesis of sugar in plants.
Sample Requirements for Disaccharides and Disaccharide Metabolites Assay
| Plant Sample Type |
Sample Volume |
Storage Conditions |
Additional Notes |
| Plant Tissues |
≥ 100 mg |
Store at -80°C |
Harvest fresh tissues or freeze immediately after collection |
| Plant Extracts |
≥ 500 µL |
Store at -80°C |
Centrifuge extracts before storage to remove debris |
| Plant Cell Cultures |
≥ 1 mL |
Store at -80°C |
Centrifuge cell cultures before storage to remove cell debris |
| Plant Sap |
≥ 100 µL |
Store at -80°C |
Collect sap using sterile techniques and avoidcontamination |
| Plant Seeds |
≥ 50 mg |
Store at -20°C |
Remove husks or shellsbefore analysis |
Each experimental treatment should have more than 6 biological replicates to ensure robust statistical analysis and reliable interpretation of results.
Case. Nutritional Component Analyses in Different Varieties of Actinidia eriantha Kiwifruit by Transcriptomic and Metabolomic Approaches
Background:
This research focuses on Actinidia eriantha, a promising germplasm resource for kiwifruit breeding. Utilizing metabolomic and transcriptomic approaches, three elite varieties were analyzed for their sugar compositions.
Gene expression patterns associated with sugar metabolism pathways were also investigated. These findings contribute to a deeper understanding of sugar metabolism in A. eriantha fruits, providing valuable insights for enhancing fruit quality and advancing molecular breeding strategies in kiwifruit cultivation.
Samples:
Three varieties 'MM-11', 'MM-13' and 'MM-16' of A. eriantha,were grafted on 8-year-old 'Jinkui' (A. deliciosa) rootstock for more than three years.
Mature fruit of the three varieties were collected immediately frozen in liquid nitrogen and stored at −80 ℃ for RNA-Seq and metabolic assays.
Technical methods procedure:
Ripe fruit samples of three A. eriantha varieties (MM-11, MM-13, and MM-16) were collected and freeze-dried. Fruit powder (100 mg) was extracted with 0.6 mL 70% aqueous methanol at 4°C overnight.
Metabolite profiling was conducted by UPLC-MS/MS analysis, using a widely targeted metabolome method.
Differentially accumulated metabolites (DAMs) were identified based on criteria including VIP ≥ 1 and fold change ≥2 or ≤0.5. Analyst 1.6.3 software was utilized for data analysis. DAMs were identified with VIP ≥ 1 and a fold change ≥2 or ≤0.5.
Results
Differential accumulated metabolites (DAMs) analysis identified 236 DAMs across the three varieties. Flavonoids and phenolic acids were predominant among the DAMs. Significant differences were observed in the content of various metabolites among the varieties.
Analysis of sugar and organic acid metabolism pathways highlighted starch degradation and sugar accumulation during fruit maturation. Ascorbic acid biosynthesis pathways were investigated, emphasizing the role of the L-galactose pathway in ascorbic acid synthesis.
The study provides insights into the nutrient profile of A. eriantha fruit and the correlation between gene expression and metabolic traits. Molecular markers associated with traits could facilitate molecular breeding approaches for kiwifruit improvement.
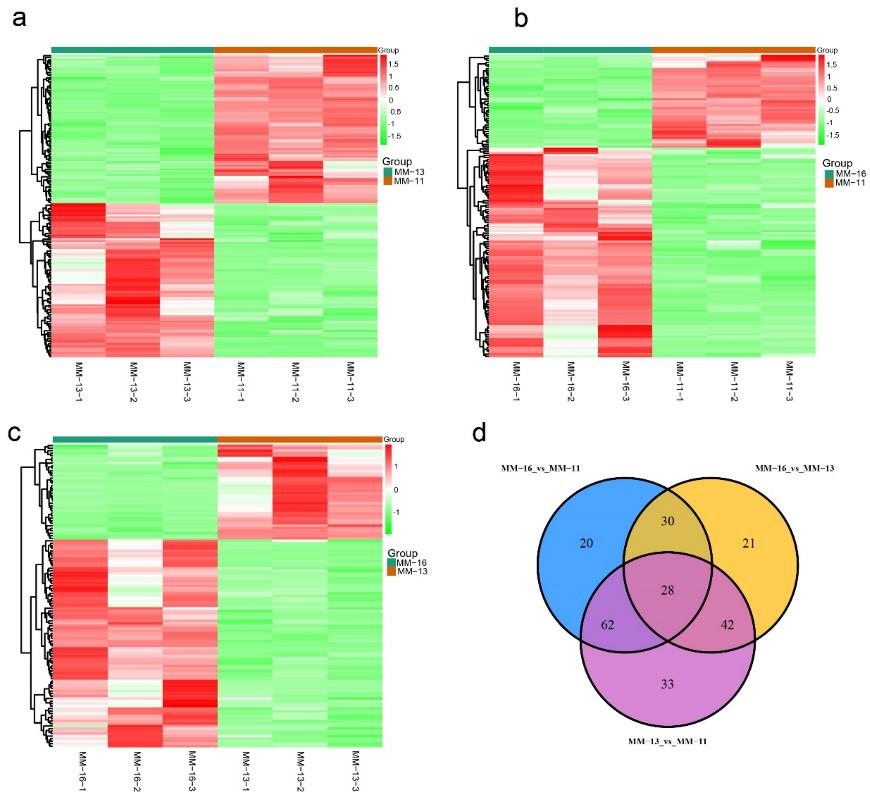 Figure 1. Differentially accumulated metabolites (DAMs) between 'MM-11', 'MM-13' and 'MM-16'. Heatmap of the DAMs in MM-11 vs. MM-13, MM-11 vs. MM-16 and MM-13 vs. MM-16 are shown in (a–c), respectively. (d) Venn diagram showing the number of DAMs in MM-11 vs. MM-13, MM-11 vs. MM-16 and MM-13 vs. MM-16.
Figure 1. Differentially accumulated metabolites (DAMs) between 'MM-11', 'MM-13' and 'MM-16'. Heatmap of the DAMs in MM-11 vs. MM-13, MM-11 vs. MM-16 and MM-13 vs. MM-16 are shown in (a–c), respectively. (d) Venn diagram showing the number of DAMs in MM-11 vs. MM-13, MM-11 vs. MM-16 and MM-13 vs. MM-16.
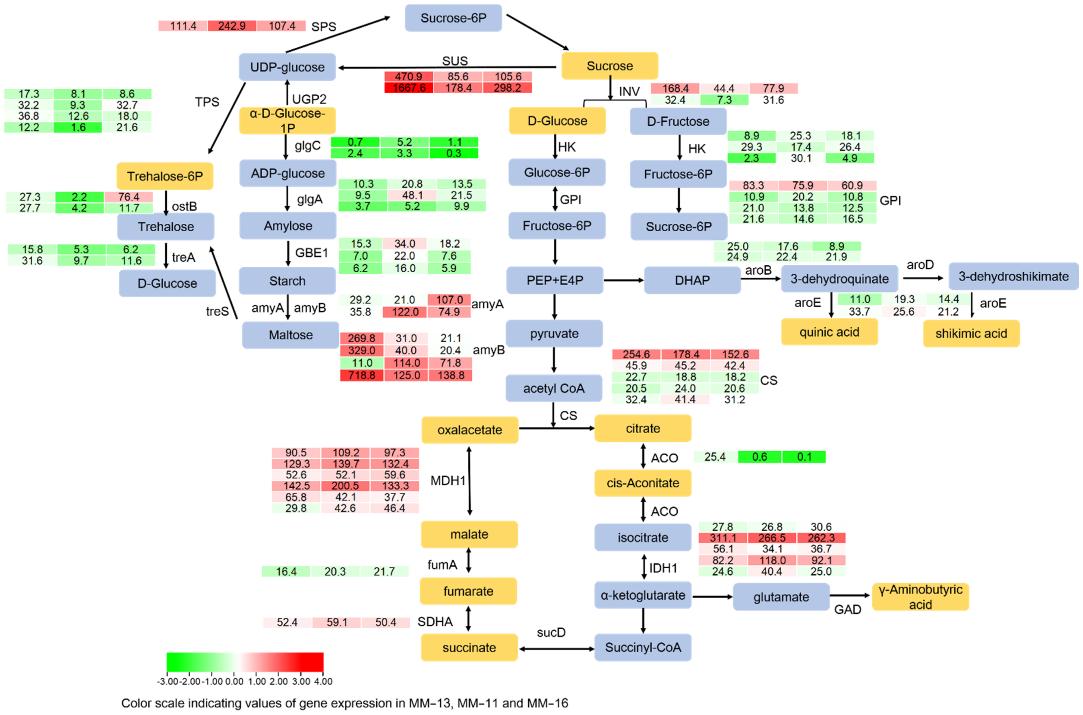 Figure 2. F Transcript profiling of genes in the starch, sucrose and organic acid biosynthetic pathways in MM-11, MM-13 and MM-16 at mature stages. The heatmap of DEGs was analyzed, and FPKM values of genes are shown.
Figure 2. F Transcript profiling of genes in the starch, sucrose and organic acid biosynthetic pathways in MM-11, MM-13 and MM-16 at mature stages. The heatmap of DEGs was analyzed, and FPKM values of genes are shown.
Reference
- Jia, H. (2022). "Nutritional Component Analyses in Different Varieties of Actinidia eriantha Kiwifruit by Transcriptomic and Metabolomic Approaches." International Journal of Molecular Sciences 23(18), 10217.



 Figure 1. Differentially accumulated metabolites (DAMs) between 'MM-11', 'MM-13' and 'MM-16'. Heatmap of the DAMs in MM-11 vs. MM-13, MM-11 vs. MM-16 and MM-13 vs. MM-16 are shown in (a–c), respectively. (d) Venn diagram showing the number of DAMs in MM-11 vs. MM-13, MM-11 vs. MM-16 and MM-13 vs. MM-16.
Figure 1. Differentially accumulated metabolites (DAMs) between 'MM-11', 'MM-13' and 'MM-16'. Heatmap of the DAMs in MM-11 vs. MM-13, MM-11 vs. MM-16 and MM-13 vs. MM-16 are shown in (a–c), respectively. (d) Venn diagram showing the number of DAMs in MM-11 vs. MM-13, MM-11 vs. MM-16 and MM-13 vs. MM-16. Figure 2. F Transcript profiling of genes in the starch, sucrose and organic acid biosynthetic pathways in MM-11, MM-13 and MM-16 at mature stages. The heatmap of DEGs was analyzed, and FPKM values of genes are shown.
Figure 2. F Transcript profiling of genes in the starch, sucrose and organic acid biosynthetic pathways in MM-11, MM-13 and MM-16 at mature stages. The heatmap of DEGs was analyzed, and FPKM values of genes are shown.

