What is Plants Untargeted Metabolomics?
Plants untargeted metabolomics is a scientific discipline that involves the comprehensive analysis of small molecules or metabolites present within a plant system. Unlike targeted metabolomics which focuses on predefined compounds, untargeted metabolomics takes a broader approach, aiming to detect and quantify as many metabolites as possible without prior knowledge of their identity. This holistic perspective provides an invaluable insight into the metabolic landscape of plants, enabling researchers to uncover novel compounds, pathways, and interactions.
Specific Projects in Plants Untargeted Metabolomics
At Creative Proteomics, we offer an extensive range of projects within the domain of plants untargeted metabolomics. Our commitment to scientific excellence is reflected in the diversity of our services, each meticulously designed to provide a comprehensive understanding of plant metabolites. Some of the specific projects we offer include:
Metabolite Profiling: A comprehensive analysis of plant samples to identify and quantify a wide spectrum of metabolites.
Pathway Discovery: Unraveling complex metabolic pathways within plants, shedding light on interconnected biochemical processes.
Metabolite Identification: Utilizing advanced mass spectrometry techniques to identify unknown metabolites and elucidate their chemical structures.
Comparative Metabolomics: Comparative analysis of metabolite profiles across different plant species or experimental conditions, facilitating insights into evolutionary and environmental adaptations.
Stress Response Analysis: Investigating how plants metabolically respond to various stressors, providing crucial insights into plant defense mechanisms.
Plants Untargeted Metabolomics Analysis Techniques
Central to the success of plants untargeted metabolomics analysis are state-of-the-art mass spectrometry techniques. Creative Proteomics employs cutting-edge instruments to unravel the intricate world of plant metabolites.
- Liquid Chromatography-Mass Spectrometry (LC-MS): LC-MS combines the separation capabilities of liquid chromatography with the sensitivity and specificity of mass spectrometry. Instruments such as Thermo Scientific Q Exactive™ HF-X enable high-resolution and accurate mass measurements.
- Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS is ideal for volatile and semi-volatile compound analysis. The Agilent 7890B GC System coupled with mass spectrometry provides unparalleled sensitivity and reproducibility.
- Nuclear Magnetic Resonance (NMR) Spectroscopy: NMR spectroscopy offers insights into the structural elucidation of metabolites. Our Bruker Avance III HD 600 MHz NMR Spectrometer ensures precise and detailed analysis.
- High-Resolution Mass Spectrometry: Instruments like Thermo Scientific Orbitrap Exploris 480 provide exceptional mass accuracy and resolution, facilitating the detection of a wide range of metabolites.
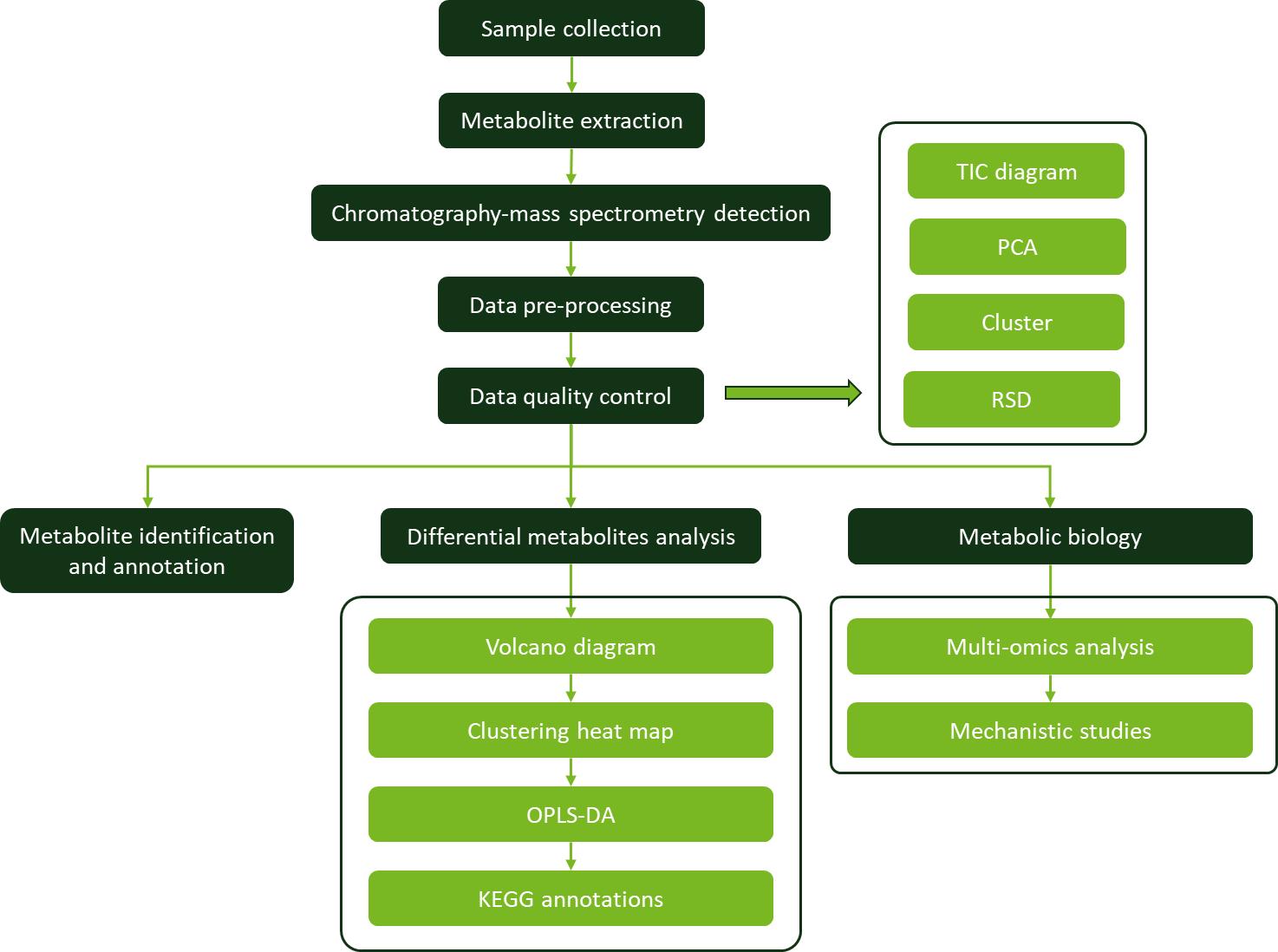 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Our Plant Metabolism Database (including but not limited to)
| Substance Category |
Compounds |
Quantity |
| Amino acids |
Threonine, serine, tyrosine, valine, glutamine, cysteine, etc. |
100+ |
| Vitamins |
Vitamin A, vitamin B12, vitamin C, vitamin D2, vitamin K1, etc. |
30+ |
| Organic acids |
Fenugreek acid, malic acid, mangiferic acid, ferulic acid citric acid, tartaric acid, etc. |
200+ |
| Carbohydrates |
Lactose, galactose, sorbitol, xylitol, mannitol, arabinose, etc. |
50+ |
| Lipids |
Linoleic acid, palmitic acid, linolenic acid, myristic acid, arachidonic acid, etc. |
100+ |
| Nucleotides |
1-methyladenosine, 5-methylcytidine, adenine, cytidine, guanine, etc. Cytisine, guanine, etc. |
100+ |
| Phenols |
Rhododendron, rhizophorin, salvinorin, curcumin, rhodiol glycosides, etc. |
80+ |
| Phenolic Acids |
Caffeic acid, catechin gallate, benzoylpaeoniflorin, salvinorin A, etc. |
60+ |
| Flavonoids |
Baicalein, baicalein, luteolin, populin, kaempferol, proanthocyanidin B1, etc. |
450+ |
| Alkaloids |
Picrasidine, Rifampicin, Betaine, Ricin, Crotonin, etc. |
370+ |
| Terpenoids |
Astragaloside I, ginsenoside, betulinol, carvone, etc. |
350+ |
| Quinones |
Dihydrotanshinone I, oryzanthin, rhodopsin, tanshinone lIIA, chrysin, etc. |
30+ |
| Steroids |
Lycorine, Ophiopogonin D, Digitalis Saponin, Allium Veratrum, Riluzin, etc. |
60+ |
| Phenylpropanoids |
Geranylgeranyl glucoside, tanshinoside, chlorogenic acid, 1-caffeoylquinic acid, etc. |
150+ |
| Others |
Bamboo rhododendron, Xanthophyllin B, Acetobenzene, Patchouli ketone, etc. |
400+ |
Applications of Plants Untargeted Metabolomics Analysis
Plant Ecology: Understanding the metabolic regulation of plants in their natural ecosystems, revealing plant responses to environmental changes and climate variations.
Agricultural Improvement: Analyzing metabolic differences among different plant varieties to provide insights for cultivating crops that are better adapted to varying soil conditions and climate environments.
Medicinal Plant Research: Uncovering bioactive compounds within medicinal plants, aiding in the development of new drugs or the enhancement of existing ones.
Food Safety and Quality: Monitoring the quality, safety, and authenticity of agricultural products by analyzing plant metabolites present in food.
Environmental Pollution Monitoring: Analyzing plant metabolites to detect pollutants or harmful substances in the environment.
Plant-Microbe Interactions: Investigating metabolic interactions between plants and microorganisms, revealing symbiotic or antagonistic mechanisms.
Genetic Studies: Revealing the relationship between genes and metabolism by comparing plant metabolomes of different genotypes, deepening our understanding of genetic mechanisms.
Grassland Ecology: Applying untargeted metabolomics to study interactions between plants and other organisms in grasslands, contributing to grassland conservation and restoration efforts.
Discovery of New Species: Applying metabolomics to unknown plant species to aid in species identification and description.
Industrial Applications: Optimizing extraction processes in the production of plant extracts using untargeted metabolomics analysis to enhance yield and quality.
Sample Requirements for Plants Untargeted Metabolomics Assay
| Sample Types |
Minimum Sample Size |
| Plant Samples |
Roots, stems and leaves, floral parts, fruits/seeds, rhizomes, buds/tender leaves, tissue sections, pollen, bark, trunk/wood, resin/gum, resin acids, seedlings/young plants, rhizosphere soil, root exudates. |
50 mg - 1 g |
Plants Targeted Metabolomics vs. Untargeted Metabolomics
| Aspects |
Plant Targeted Metabolomics |
Plant Untargeted Metabolomics |
| Purpose |
Precisely quantify specific metabolites of interest |
Explore the complete metabolite landscape without bias |
| Sample Preparation |
Requires prior knowledge of target compounds |
No prior knowledge needed, allows for unbiased analysis |
| Analysis Approach |
Employ specific assays for target metabolites |
Utilize high-resolution techniques for broad spectrum analysis |
| Data Analysis |
Focuses on quantifying pre-defined metabolites |
Involves complex data mining for holistic metabolite profiling |
| Metabolite Identification |
Based on comparison with known standards |
Relies on spectral matching and databases for compound identification |
| Key Advantages |
Accurate quantification, targeted insight |
Discovery of novel compounds and broader metabolic insights |
| Application Examples |
Investigate plant responses to specific stressors |
Uncover diverse metabolic changes under varying conditions |
Case 1. Untargeted Metabolomics Reveals Defense-Related Metabolic Pathways in Soybean Plants During Asian Soybean Rust Infection
Background:
Plant-pathogen interactions are critical determinants of agricultural productivity. Among these, Asian soybean rust (ASR) caused by Phakopsora pachyrhizi is a major threat to soybean crops. Investigating the metabolic changes in response to ASR is essential for developing effective disease management strategies. This study focused on comparing metabolite profiles of soybean plants inoculated with P. pachyrhizi to understand the plant's defense mechanisms. Utilizing advanced techniques like UHPLC-ESI-MS/MS and molecular networking, the research sheds light on pathways involving fatty acids, phenylpropanoids, flavonoids, isoflavonoids, coumarins, and terpenes that contribute to soybean's resistance against ASR.
Samples:
In this study, soybean plants (Glycine max) of a resistant cultivar (PI567025A, Rpp3 gene) were investigated. The focus was on comparing the metabolite profiles of control plants and those inoculated with Phakopsora pachyrhizi spores, the causal agent of ASR.
Methods:
The researchers employed an untargeted metabolomics approach, utilizing ultra-high-performance liquid chromatography (UHPLC) coupled with hybrid quadrupole time-of-flight high-resolution mass spectrometry (MS/MS). The workflow involved metabolite extraction from plant leaves, followed by data processing and analysis.
Metabolite Extraction: Soybean leaves were macerated and extracted using a chloroform/methanol/water solvent system. This method preserved metabolites and allowed for subsequent analysis.
UHPLC-ESI-MS/MS Analysis: The extracted metabolites were subjected to UHPLC separation using a C18 column with a binary solvent system. The mass spectrometer, equipped with an electrospray ionization source, facilitated the detection and fragmentation of metabolites. Data collection covered a wide m/z range, enabling the identification of different metabolites.
Data Processing and Analysis: The raw data underwent preprocessing using the XCMS software for feature detection, retention time correction, alignment, and intensity. MetaboAnalyst 3.0 was utilized for multivariate analysis, including Principal Component Analysis (PCA) and Partial Least Squares Discriminant Analysis (PLS-DA), to assess metabolic differences between control and inoculated plants.
Molecular Networking: A molecular network was constructed using the Global Natural Products Social Molecular Networking (GNPS) platform. This facilitated the grouping of related compounds based on their fragmentation patterns, aiding in chemical classification and identification.
Results
The results revealed significant changes in the metabolite profiles of soybean plants inoculated with P. pachyrhizi spores compared to control plants. Notably, several metabolic pathways associated with plant defense responses were activated:
- Polyunsaturated fatty acids were identified, suggesting involvement in the synthesis of jasmonic acid, a key defense hormone.
- Phenylpropanoid pathway metabolites, such as flavonoids and isoflavonoids, were up-regulated, indicating their role in defense against ASR.
- Coumarins, known for their antimicrobial properties, were detected and potentially contributed to defense mechanisms.
- Triterpenic saponins, categorized as phytoanticipins, showed a presence in the metabolome, potentially playing a role in defense.
The study underscored the value of untargeted metabolomics in deciphering complex plant-pathogen interactions and provided insights into the metabolic responses of soybean plants to ASR infection. The detailed analysis of metabolite profiles contributes to the broader understanding of plant defense strategies and offers avenues for further research in crop protection and resistance enhancement.
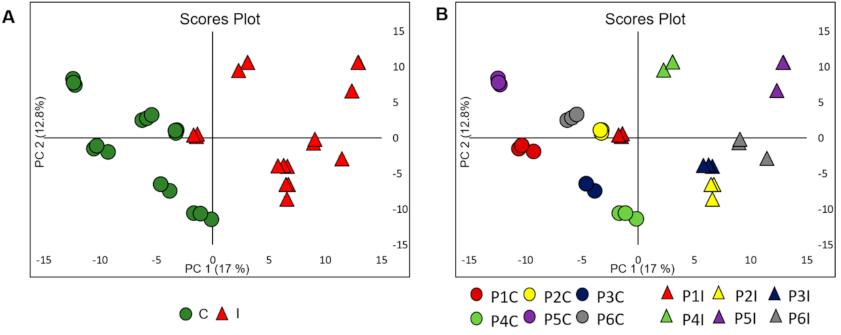 Unsupervised chemometric modeling (UHPLC-ESI-(+)-MS/MS data)
Unsupervised chemometric modeling (UHPLC-ESI-(+)-MS/MS data)
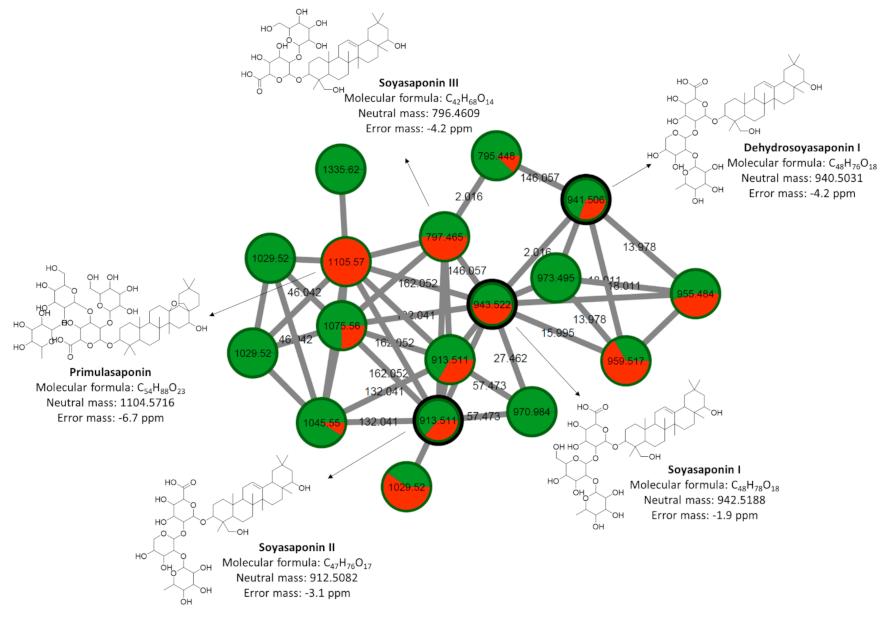 Cluster of terpenes containing triterpene saponins putatively characterized by molecular network obtained from MS/MS data from control plants and inoculated plants with P. pachyrhizi.
Cluster of terpenes containing triterpene saponins putatively characterized by molecular network obtained from MS/MS data from control plants and inoculated plants with P. pachyrhizi.
Reference
- Silva, Evandro, et al. "Untargeted metabolomics analysis by UHPLC-MS/MS of soybean plant in a compatible response to Phakopsora pachyrhizi infection." Metabolites 11.3 (2021): 179.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
 Unsupervised chemometric modeling (UHPLC-ESI-(+)-MS/MS data)
Unsupervised chemometric modeling (UHPLC-ESI-(+)-MS/MS data) Cluster of terpenes containing triterpene saponins putatively characterized by molecular network obtained from MS/MS data from control plants and inoculated plants with P. pachyrhizi.
Cluster of terpenes containing triterpene saponins putatively characterized by molecular network obtained from MS/MS data from control plants and inoculated plants with P. pachyrhizi.

