What is Arginine Metabolism?
Arginine (Arg) is a crucial amino acid in plants, serving as a precursor for compounds such as nitric oxide (NO), polyamines, and proline (Pro), which are essential for growth, development, and stress responses. It is metabolized via the ornithine and arginine decarboxylase pathways, leading to the production of important molecules like putrescine (Put) and NO. These pathways and their products play significant roles in stress adaptation by enhancing tolerance to environmental stresses like salinity and drought.
Arginine metabolism supports the development of leaves, stems, roots, and shoots, and integrates into the hormonal regulation of plant growth, contributing to overall plant health and vigor. Here at Creative Proteomics, we provide comprehensive services for arginine metabolism analysis. These services are meticulously crafted to enrich your comprehension and optimization of plant growth dynamics and resilience to stressors.
Arginine Metabolism Analysis Service by Creative Proteomics
Arginine Metabolism Metabolite Profiling: Utilizing cutting-edge analytical methodologies, we conduct comprehensive profiling of metabolites implicated in arginine pathways. This encompasses proline, polyamines (putrescine, spermidine, spermine), and NO, furnishing detailed insights into their distribution and dynamics.
Arginine Metabolism Enzyme Activity Assessment: Our suite of services involves precise quantification of pivotal enzyme activities like arginase (ARG), arginine decarboxylase (ADC), ornithine (ODC), and nitric oxide synthase (NOS). This facilitates a thorough grasp of their functionalities and regulatory mechanisms, which is crucial for optimizing metabolic pathways.
Arginine Metabolism Stress Response Evaluation: Through meticulous examination, our team delves into the intricate dynamics of arginine metabolism under diverse stress conditions. By meticulously studying the responses of arginine pathways to stressors such as salinity, drought, and temperature fluctuations, we offer invaluable insights into the mechanisms bolstering plant resilience and survival strategies.
Techniques and Instrumentation for Arginine Metabolism Analysis
Liquid Chromatography (LC): Central to our service is the utilization of liquid chromatography (LC), which enables the separation and purification of arginine and its metabolites before mass spectrometry analysis. Our LC systems (Vanquish H) are renowned for their superior chromatographic resolution and efficiency.
Mass Spectrometry (MS): The high-resolution MS instrument (Thermo Q-Exactive) allows for the precise detection and quantification of arginine and its metabolites. With exceptional mass accuracy and resolution, our MS platforms enable accurate characterization and quantification of arginine-related compounds across a wide range of plant samples.
LC-MS/MS Method Establishment and Optimization: We establish precise quantification methods using reference standards to ensure accurate determination of arginine and related compounds. Calibration procedures guarantee linearity and reproducibility, facilitating accurate measurement across concentrations. Multiple Reaction Monitoring (MRM) enhances detection efficiency by minimizing interference and optimizing analysis sensitivity.
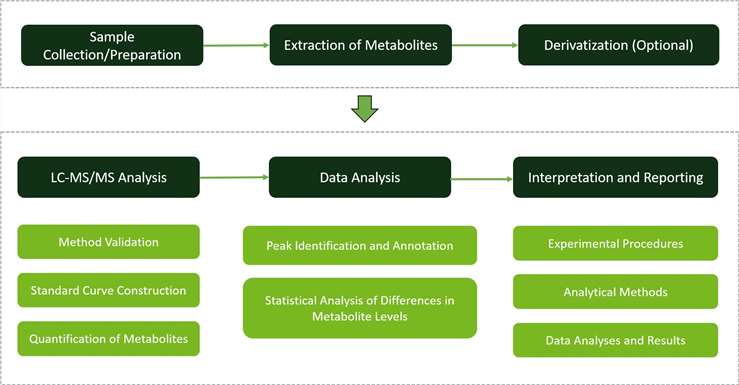
Why Choose Us?
- Comprehensive Analysis: At Creative Proteomics, we adopt a holistic approach to investigating arginine metabolism, encompassing diverse facets ranging from metabolic profiling to enzyme activity assays. This meticulous examination fosters a detailed understanding of how arginine and its derivatives influence plant growth, development, and stress responses.
- Cutting-edge Techniques: We employ advanced methods for the precise quantification and identification of arginine and its metabolites. The employment of LC-MS/MS technology ensures exceptional chromatographic resolution and unparalleled sensitivity and specificity. Therefore, researchers can effectively address challenges posed by samples with high impurity levels or low concentrations of target analytes.
- Customized Solutions: Recognizing the unique requirements of each research project, Creative Proteomics offers tailored experimental designs and analytical methods. Solutions are designed to optimize protocols and align with specific research objectives. Whether it entails adjusting chromatographic conditions or MS settings, our adaptable approaches are tailored precisely to meet your research needs.
- Experienced Team: Seasoned scientists proficient in plant metabolism analysis allow us to deliver high-quality results and provide valuable insights into the intricacies of arginine metabolism. We are committed to unraveling the complexities of arginine metabolism, fostering a deeper understanding of its significance in plants.
- In-house Database: We have developed an extensive in-house database by integrating commercial information libraries with our proprietary data. This repository encompasses a wide array of standard compounds and reference materials, facilitating the accurate qualification and quantification of numerous arginine metabolites.
Applications of Arginine Metabolism Analysis
Plant Physiology Research: Creative Proteomics aids researchers in unraveling the role of arginine metabolism in plant growth, development, and stress responses. By understanding these mechanisms, novel strategies can be designed to enhance crop productivity or stress tolerance.
Crop Improvement Programs: Insights gained from our analysis can inform breeding programs aimed at developing cultivars with improved arginine metabolism traits. This contributes to the development of resilient and high-yielding crop varieties suited for diverse environmental conditions.
Biotechnological Applications: Creative Proteomics supports biotechnological endeavors focused on engineering metabolic pathways for enhanced arginine production or stress tolerance in plants. These applications have implications in the production of biofuels, pharmaceuticals, and nutraceuticals.
Environmental Studies: Understanding arginine metabolism in plants is crucial for assessing their role in environmental processes such as nitrogen cycling and pollutant remediation. Our service facilitates research in these areas, contributing to environmental conservation efforts.
Sample Requirements for Arginine Metabolism Assay
| Plant Sample Type |
Minimum Quantity Required (fresh weight) |
Storage Conditions |
Additional Notes |
| Roots, Stems, Leaves, Fruits |
>200 mg |
Quick freeze in liquid nitrogen, Store at -80°C |
- |
| Seeds |
>200 mg |
Include seeds if applicable |
Each experimental treatment should have more than 6 biological replicates to ensure robust statistical analysis and reliable interpretation of results.
For other sample types not listed above, such as flowers or whole plants, please consult our technical support or sales team for specific requirements and recommendations.
Case. Transcriptomics and Metabolomics Changes Triggered by Exogenous 6-Benzylaminopurine in Relieving Epicotyl Dormancy of Polygonatum cyrtonema Hua Seeds
Background:
Polygonatum cyrtonema Hua, a valuable medicinal plant, faces cultivation challenges due to seed epicotyl dormancy, requiring two winters for germination.
Cytokinins like 6-benzylaminopurine (6-BA) promote dormancy release, but the molecular mechanisms in P. cyrtonema are unclear. This study uses transcriptomics and metabolomics to elucidate these mechanisms, aiding large-scale cultivation and commercial use.
Samples:
After 60 days in wet sand at 4°C, Polygonatum cyrtonema seeds were sown in soil and grown in a climate chamber (25°C, dark).
After 30 days, 200 mg/L 6-BA was applied to seeds with 0.1–0.2 cm epicotyl buds, with water as the control.
Samples were collected at various intervals, snap-frozen in liquid nitrogen, and stored at -80°C. Germination rate, root length, and bud length were measured every 5 days.
Technical methods procedure:
High-throughput mRNA sequencing was performed on seeds at three stages (12 h, 7 days, and 28 days post-6-BA treatment). RNA was extracted using the CTAB method, sequenced using the Illumina NovaSeq 6000 platform, and analyzed to identify differentially expressed genes (DEGs).
RNA-seq reads were mapped and quantified using Bowtie2 and RSEM software. DEGs were identified using DESeq with criteria |log2 (fold-change)| ≥ 1 and adjusted p-value < 0.05. Gene Ontology (GO) and KEGG pathway enrichment analyses were performed.
Metabolic profiling was conducted simultaneously with transcriptomic sampling. Freeze-dried samples were processed and analyzed using UHPLC-MS. Data were processed and annotated using SCIEX Analyst Workstation and in-house R programs.
Correlation analysis of DEGs and differentially expressed metabolites (DEMs) was performed, and networks were visualized using Cytoscape. Integrated analyses, including significance and PCA, were conducted using R software. Visualization was done using GraphPad Prism and Adobe Photoshop.
Results
Germination rates increased over time, peaking at 60% by day 30. Application of 6-BA increased emergence rates, particularly at concentrations between 100-300 mg/L. Seeds treated with 6-BA showed faster emergence compared to untreated seeds.
Seed samples were collected at various stages post-6-BA treatment. Significant changes were observed in flavonoid biosynthesis, arginine/proline metabolism, and zeatin biosynthesis pathways. Correlation between gene expression and metabolite levels highlighted key pathways influencing dormancy release.
Differentially expressed genes were identified across stages, particularly related to carbohydrate metabolism, photosynthesis, and hormonal pathways. Gene functions were associated with dormancy release processes, including flavonoid biosynthesis and hormone signaling.
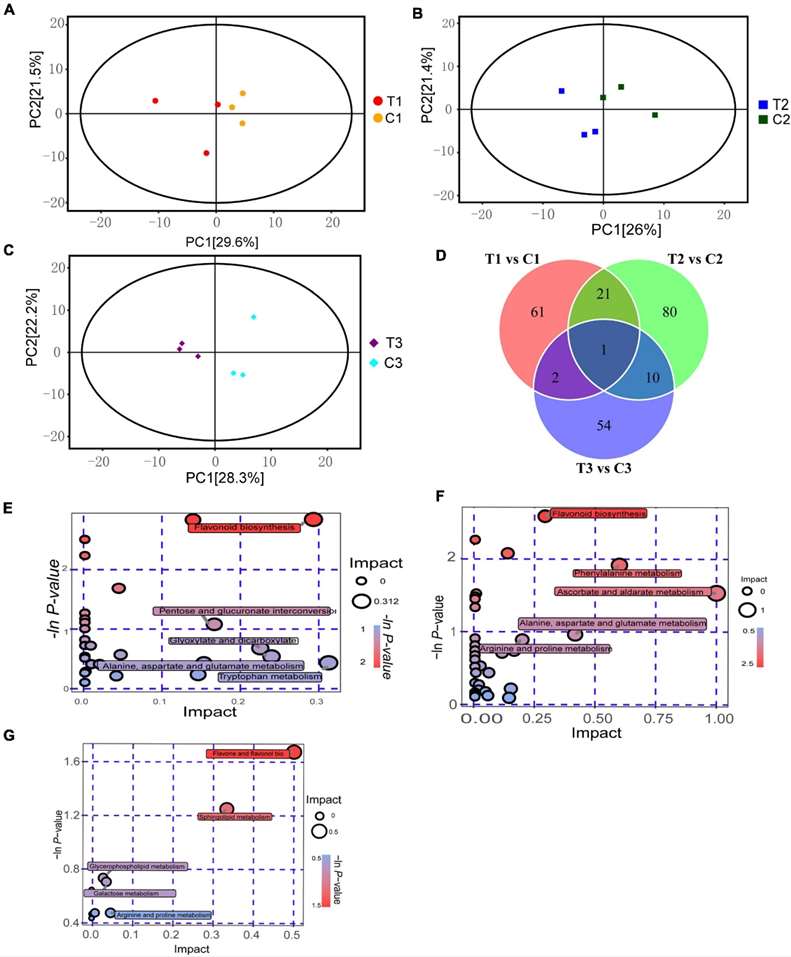 Fig 1. Metabolomics analysis on dormancy release of epicotyl in P. cyrtonema seeds. (A) PCA analysis of T1 vs. C1. (B) PCA analysis of T2 vs. C2. (C) PCA analysis of T3 vs. C3. (D) Venn diagram of metabolite distributions across different stages. (E) KEGG analysis of T1 vs. C1. (F) KEGG analysis of T2 vs. C2. (G) KEGG analysis of T3 vs. C3.
Fig 1. Metabolomics analysis on dormancy release of epicotyl in P. cyrtonema seeds. (A) PCA analysis of T1 vs. C1. (B) PCA analysis of T2 vs. C2. (C) PCA analysis of T3 vs. C3. (D) Venn diagram of metabolite distributions across different stages. (E) KEGG analysis of T1 vs. C1. (F) KEGG analysis of T2 vs. C2. (G) KEGG analysis of T3 vs. C3.
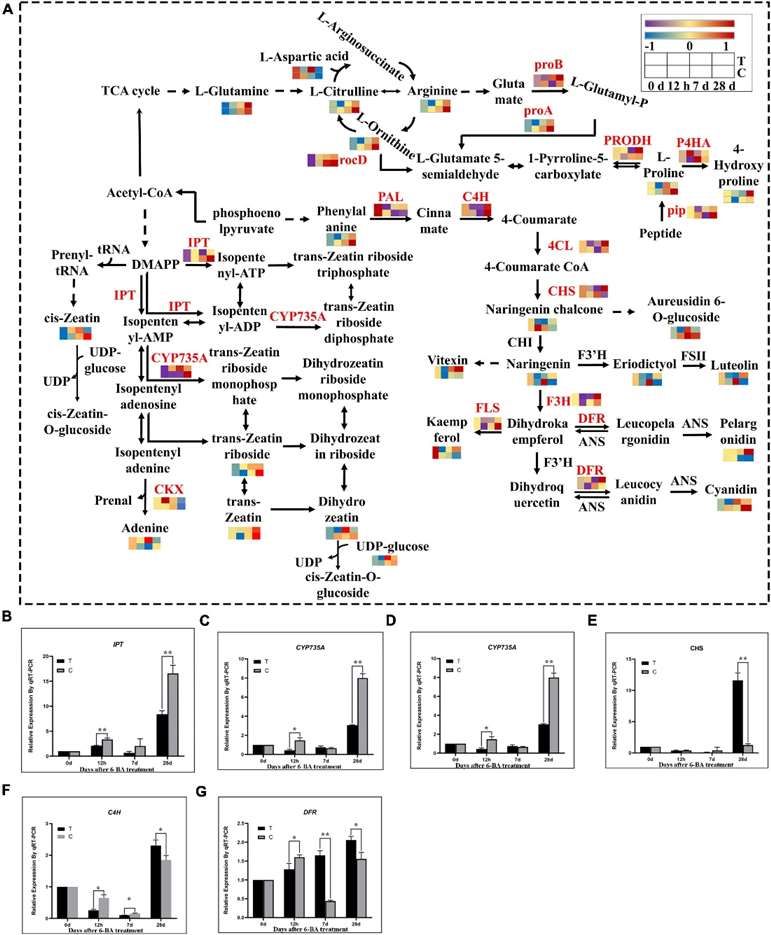 Fig 2. Integrated transcriptomics and metabolomics analysis of flavonoid biosynthesis, arginine and proline metabolism, and zeatin biosynthetic pathways. (A) The DEGs and DEMs involved in the flavonoid biosynthesis, arginine and proline metabolism, and zeatin biosynthetic pathway at different stages. T, Treatment group; C, Control group. (B–G) Verification of related genes in transcriptome by qRT-PCR. IPT, isopentenyl transferases gene; CYP735A, cytokininhydroxylase gene; CKX, cytokinin degrading enzyme gene; CHS, chalcone synthase gene; C4H, cinnamate-4-hydroxylase; DFR, dihydroflavonol 4-reductase.
Fig 2. Integrated transcriptomics and metabolomics analysis of flavonoid biosynthesis, arginine and proline metabolism, and zeatin biosynthetic pathways. (A) The DEGs and DEMs involved in the flavonoid biosynthesis, arginine and proline metabolism, and zeatin biosynthetic pathway at different stages. T, Treatment group; C, Control group. (B–G) Verification of related genes in transcriptome by qRT-PCR. IPT, isopentenyl transferases gene; CYP735A, cytokininhydroxylase gene; CKX, cytokinin degrading enzyme gene; CHS, chalcone synthase gene; C4H, cinnamate-4-hydroxylase; DFR, dihydroflavonol 4-reductase.
Reference
- Zhang, W. (2022). "Transcriptomics and metabolomics changes triggered by exogenous 6-benzylaminopurine in relieving epicotyl dormancy of Polygonatum cyrtonema Hua seeds." Frontiers in Plant Science 13, 961899.



 Fig 1. Metabolomics analysis on dormancy release of epicotyl in P. cyrtonema seeds. (A) PCA analysis of T1 vs. C1. (B) PCA analysis of T2 vs. C2. (C) PCA analysis of T3 vs. C3. (D) Venn diagram of metabolite distributions across different stages. (E) KEGG analysis of T1 vs. C1. (F) KEGG analysis of T2 vs. C2. (G) KEGG analysis of T3 vs. C3.
Fig 1. Metabolomics analysis on dormancy release of epicotyl in P. cyrtonema seeds. (A) PCA analysis of T1 vs. C1. (B) PCA analysis of T2 vs. C2. (C) PCA analysis of T3 vs. C3. (D) Venn diagram of metabolite distributions across different stages. (E) KEGG analysis of T1 vs. C1. (F) KEGG analysis of T2 vs. C2. (G) KEGG analysis of T3 vs. C3. Fig 2. Integrated transcriptomics and metabolomics analysis of flavonoid biosynthesis, arginine and proline metabolism, and zeatin biosynthetic pathways. (A) The DEGs and DEMs involved in the flavonoid biosynthesis, arginine and proline metabolism, and zeatin biosynthetic pathway at different stages. T, Treatment group; C, Control group. (B–G) Verification of related genes in transcriptome by qRT-PCR. IPT, isopentenyl transferases gene; CYP735A, cytokininhydroxylase gene; CKX, cytokinin degrading enzyme gene; CHS, chalcone synthase gene; C4H, cinnamate-4-hydroxylase; DFR, dihydroflavonol 4-reductase.
Fig 2. Integrated transcriptomics and metabolomics analysis of flavonoid biosynthesis, arginine and proline metabolism, and zeatin biosynthetic pathways. (A) The DEGs and DEMs involved in the flavonoid biosynthesis, arginine and proline metabolism, and zeatin biosynthetic pathway at different stages. T, Treatment group; C, Control group. (B–G) Verification of related genes in transcriptome by qRT-PCR. IPT, isopentenyl transferases gene; CYP735A, cytokininhydroxylase gene; CKX, cytokinin degrading enzyme gene; CHS, chalcone synthase gene; C4H, cinnamate-4-hydroxylase; DFR, dihydroflavonol 4-reductase.

