Metabolomics is a powerful approach in biological research that involves the comprehensive analysis of small molecule metabolites within a biological system. When applied to Malus domestica, commonly known as the apple tree, metabolomics allows researchers to gain valuable insights into the metabolic processes, biochemical pathways, and responses to various factors that influence apple growth, development, and overall physiology.
The metabolome of Malus domestica comprises a diverse range of metabolites, including carbohydrates, organic acids, lipids, amino acids, secondary metabolites, and more. These metabolites are involved in essential functions such as energy production, growth, defense mechanisms, and signaling. Analyzing the metabolome provides a snapshot of the physiological state of the apple tree, capturing the dynamic changes that occur in response to genetic, environmental, and developmental cues.
Metabolomics Analysis Methods We Offer
Untargeted Metabolite Profiling by Mass Spectrometry (MS): Leveraging state-of-the-art MS platforms, we conduct untargeted analyses to comprehensively detect and quantify an extensive array of metabolites in Malus domestica. This includes carbohydrates, organic acids, amino acids, lipids, and related bioactive compounds.
Targeted Metabolite Quantification: Our targeted approach involves quantifying specific classes of metabolites, such as phytohormones and specialized metabolites. By employing quantitative MS techniques, we discern subtle changes in these metabolites within the context of Malus domestica.
Metabolic Pathway Analysis and Integration: Integrating multi-omics datasets, we engage in metabolic pathway reconstruction. This integrative approach sheds light on the interplay between genes, enzymes, and metabolites, contributing to a comprehensive understanding of metabolic network dynamics.
Specific Malus domestica Analysis Projects We Provide
Temporal Profiling of Growth and Development: Profiling metabolite dynamics across different growth and developmental stages of apple trees reveals intricate metabolic shifts that underlie physiological changes during various life stages.
Plant Hormone Dynamics: Investigating alterations in plant hormone metabolites enables an in-depth examination of regulatory mechanisms governing growth, signaling, and response pathways in Malus domestica.
Environmental Response Assessment: By scrutinizing alterations in metabolite patterns triggered by external environmental factors, we offer insights into the metabolic strategies employed by Malus domestica to adapt and respond to its surroundings.
Phytochemical Defense Mechanisms: Detection and analysis of defense-related metabolites in response to biotic stressors elucidate the intricate biochemical defense mechanisms of Malus domestica against pests and pathogens.
Transcript-Metabolite Correlation Studies: Integration of metabolomics and transcriptomics data unveils correlations between gene expression and metabolite profiles, unraveling the regulatory mechanisms influencing metabolic shifts.
Climate Adaptation Dynamics: Comparative analysis of metabolite profiles across different climatic conditions informs our understanding of how Malus domestica adapts its metabolic strategies in response to varying environments.
Genotypic Metabolic Variation: Exploring metabolite variations among diverse apple varieties provides valuable insights into the genetic underpinnings of distinct metabolic profiles, thereby informing breeding strategies and genetic investigations.
Malus domestica Metabolomics Analysis Techniques
Gas Chromatography-Mass Spectrometry (GC-MS):
- Instrument Model: Agilent 7890A GC coupled with Agilent 5975C MSD
- Advantages: Ideal for volatile and thermally stable metabolites. Provides high separation efficiency and quantification, suitable for targeted analysis.
Liquid Chromatography-Mass Spectrometry (LC-MS):
- Instrument Model: Thermo Fisher Q Exactive HF-X
- Advantages: Versatile for a wide range of metabolites. High-resolution MS enables accurate mass measurement and both targeted and untargeted analysis.
Direct Infusion Mass Spectrometry (DIMS):
- Instrument Model: Waters SYNAPT G2-Si
- Advantages: Rapid screening and identification of metabolites without separation, suitable for high-throughput analysis.
Ion Mobility Mass Spectrometry (IM-MS):
- Instrument Model: Waters SELECT SERIES Cyclic IMS
- Advantages: Separates ions based on size, shape, and charge, providing structural information. Useful for isomer separation and structural elucidation.
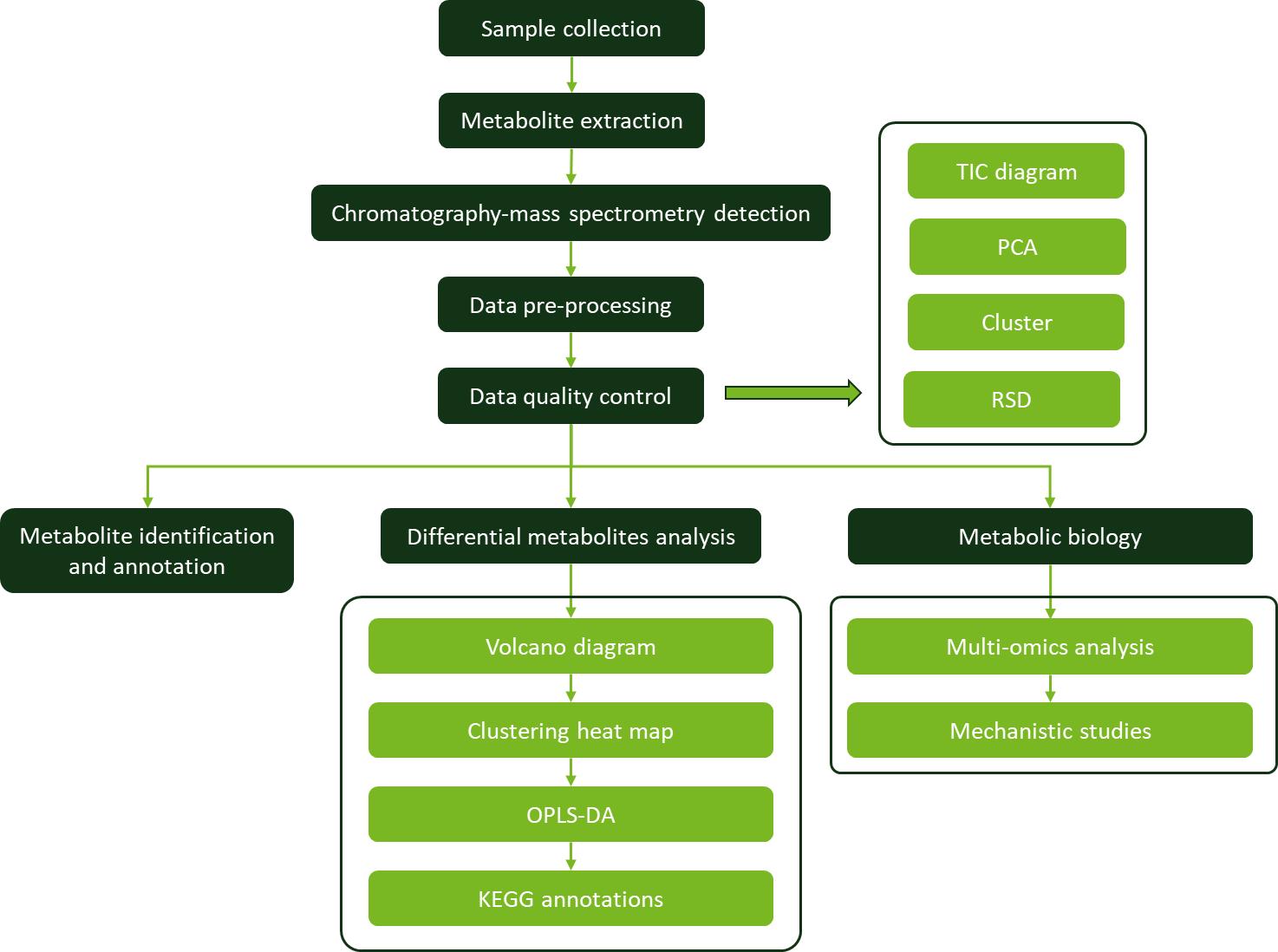 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Sample Requirements for Malus domestica Metabolomics
| Aspect |
Requirement |
| Sample Type |
Fresh apple peel, flesh, seeds, or juice. |
| Sample Size |
100 - 200 mg for solid tissues. 1 - 2 mL for liquid samples (juice). |
| Sample Collection |
Samples should be collected from various apple cultivars or conditions, all at a consistent developmental stage, and multiple time points, while also including biological replicates. |
| Sample Storage |
Samples should be rapidly frozen in liquid nitrogen upon collection and stored at -80°C to ensure preservation of metabolite stability and consistency. |
| Derivatization (if needed) |
For sugars: Silylation; For organic acids: Methylation |
| Quality Control Samples |
Include pooled QC samples covering all sample types |
| Replicates |
Include biological replicates for each condition |
| Documentation |
Detailed records of sample collection, preparation procedures, and any variations should be maintained, while considering the use of standardized protocols for consistent data quality. |
Case 1. Metabolic and Transcriptomic Profiling of 'Pinova' Apple Fruit Development
Background:
Understanding the metabolic changes during fruit development and ripening is crucial for improving fruit quality. This study aimed to comprehensively analyze the metabolomic profiles of "Pinova" apple fruits at different developmental stages.
Samples:
The study utilized "Pinova" apple fruits from four developmental stages: PS1 (27 DAA, cell division), PS2 (84 DAA, cell expansion and starch degradation), PS3 (125 DAA, preripening), and PS4 (165 DAA, full ripening). Fruit samples were collected from healthy trees and pooled to create replicates for analysis.
Methods:
Sample Preparation and Extraction:
The study focused on "Pinova" apple trees at four developmental stages: PS1, PS2, PS3, and PS4. Collected fruits were carefully freeze-dried and ground into a fine powder using a grinding miller. For metabolomics analysis, a portion of this powder was dissolved in an extraction buffer containing pure methanol with added lidocaine for lipid-soluble metabolites. Additionally, a trace amount of 2-Chloro-phenylalanine was included. This mixture was left to extract overnight in a cool environment, followed by centrifugation to obtain the extract. A Carbon-GCB SPE Cartridge was used to absorb the supernatant, and a 0.22-µm filter was applied before LC-MS/MS analysis. Another technique was used for measuring fructose content, involving extraction with methanol and MTBE, followed by a derivatization reaction.
LC-ESI-MS/MS Metabolomics Analysis:
The analysis was performed using a combination of a Shim-pack UFLC system and an Applied Biosystems 4500 Q TRAP MS/MS system. This LC-MS setup utilized an electrospray ionization (ESI) source operating in the positive-ion mode. The separation of compounds was achieved through a C18 column, and quantification relied on Multiple Reaction Monitoring (MRM) scans. These scans allowed the researchers to observe the transitions of specific ions. Furthermore, collision-induced dissociation (CID) was employed to fragment ions for structural information. The acquired data were processed and integrated using software.
Metabolomics Data Processing and Analysis:
To ensure the consistency of the analysis, a quality control mix sample was created by pooling extracts from all samples. The raw LC-MS data, comprising information about 462 metabolites, underwent normalization. The data were then used to create visual representations like heat maps and hierarchical analyses to discern trends in metabolite variations. Significantly changed metabolites were identified using the OPLS-DA model, and Principal Component Analysis (PCA) provided insights into the overall data structure. Pathway assignments were carried out using KEGG analysis.
RNA Extraction and Transcriptome Analysis:
For the transcriptomic analysis, total RNA was extracted from the apple samples and assessed for quality. The process involved removing ribosomal RNA using specialized kits. Libraries for sequencing were prepared from the remaining RNA molecules, and high-throughput sequencing was conducted using the Illumina HiSeq 4000 platform. The resulting reads were aligned to the reference genome, and gene expression levels were quantified. Differentially expressed genes were identified based on statistical analysis. Pathway analysis was performed using specialized tools.
Results
Metabolic Changes: Metabolic variations were observed across developmental stages. Sugar (Fru, Suc, Glc) accumulation increased, while organic acids (malic acid, succinic acid, fumaric acid) and flavonoids decreased.
Flavonoid Regulation: The downregulation of flavonoid pathway genes led to a rapid decrease in flavonoid content during early stages.
Primary Metabolism Activation: Genes involved in primary metabolism (TCA cycle, amino acid metabolism) were upregulated, even though corresponding metabolite levels decreased.
Metabolic Connectivity: Correlation analysis revealed close associations between primary and secondary metabolism, providing insights into the metabolic basis of apple quality traits.
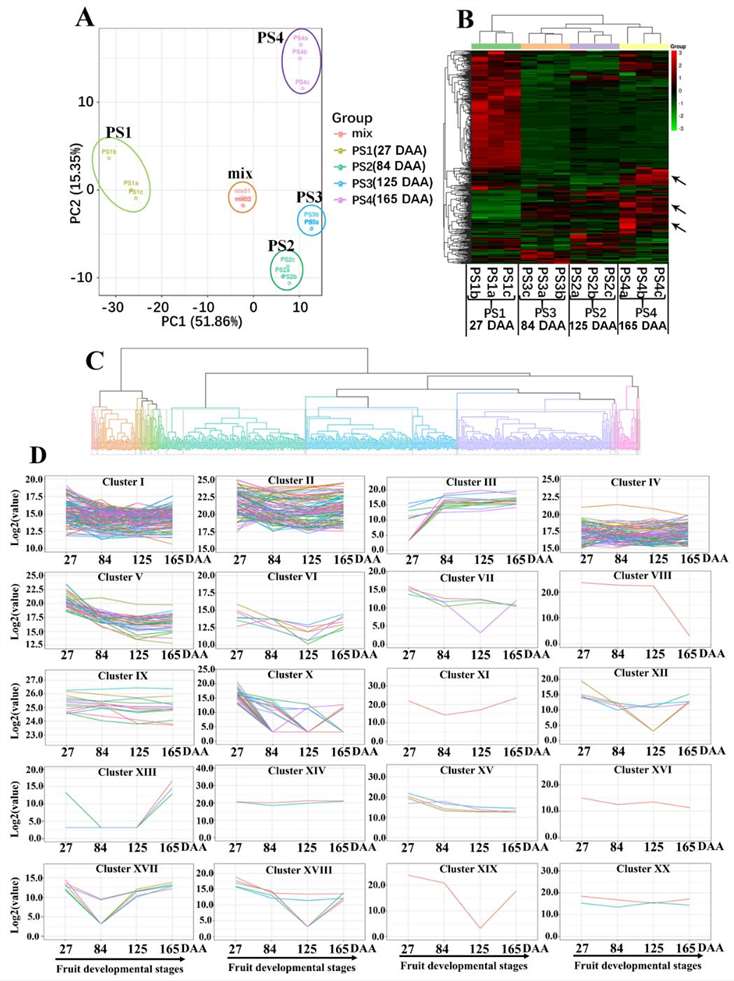 Dynamic metabolome of apple development and ripening.
Dynamic metabolome of apple development and ripening.
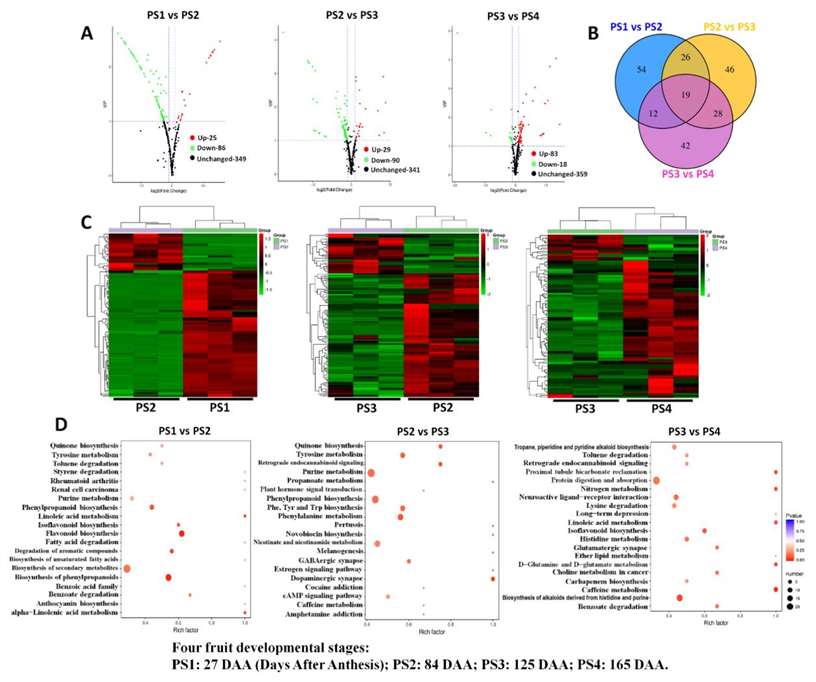 Differentially accumulated metabolites among different fruit-development stages
Differentially accumulated metabolites among different fruit-development stages
Reference
- Xu, Jidi, et al. "Integrative analyses of widely targeted metabolic profiling and transcriptome data reveals molecular insight into metabolomic variations during apple (Malus domestica) fruit development and ripening." International Journal of Molecular Sciences 21.13 (2020): 4797.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service Dynamic metabolome of apple development and ripening.
Dynamic metabolome of apple development and ripening. Differentially accumulated metabolites among different fruit-development stages
Differentially accumulated metabolites among different fruit-development stages

