The plant-environment interaction metabolome allows the analysis of small molecules or metabolites that are involved in dynamic biochemical interactions between plants and their surroundings. It encompasses the complex network of metabolic reactions and responses that occur when plants interact with a variety of environmental factors, such as soil conditions, temperature, light, moisture, biotic stressors (e.g., pests and pathogens) and abiotic stressors (e.g., drought) or nutrient availability).
Essentially, the metabolome of a plant's interactions with its environment is a snapshot of the chemical communication and adaptive processes that occur in a plant in response to its surroundings. It encompasses a wide range of metabolites, including primary metabolites (e.g., sugars, amino acids) and secondary metabolites (e.g., phytochemicals, terpenoids, and phenolic compounds), all of which play an important role in mediating the plant's response to environmental signals.
Metabolomic technologies can provide insights into how plants perceive and respond to their environment, enabling you to study the biochemical mechanisms behind plant growth, stress tolerance, defense responses, and adaptation strategies. By analyzing changes in the metabolome of plants interacting with their environment, we can better understand how plants thrive, survive and interact with their ecosystems.
Plant-Environment Interaction Metabolomics Solutions
 Metabolic Interactions Between Plants and Soil Microorganisms
Metabolic Interactions Between Plants and Soil Microorganisms
Beneath the Earth's surface lies a thriving ecosystem teeming with microorganisms crucial for plant health and growth. Soil is not just a matrix; it's an intricate network where plants engage in metabolic dialogues with microorganisms. Our cutting-edge mass spectrometry-based techniques, including liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS), are the keys to deciphering these hidden interactions. We unveil the secrets of nutrient cycling, disease suppression, and plant growth promotion.
- Nutrient Exchange: Metabolomics allows us to identify and quantify the metabolites involved in nutrient exchange between plants and soil microorganisms. This includes understanding how microorganisms release essential nutrients like nitrogen, phosphorus, and iron, and how plants take up these nutrients through metabolic pathways.
- Disease Suppression: We investigate the production of secondary metabolites by both plants and soil microorganisms. These secondary metabolites often have antimicrobial properties, shedding light on disease suppression mechanisms in the rhizosphere.
- Plant Growth Promotion: Metabolomics helps uncover the metabolic pathways involved in plant growth promotion by soil microorganisms. This includes the production of growth-promoting hormones, such as auxins and cytokinins, and the enhancement of nutrient availability.
 Metabolic Regulation in Response to Pests and Pathogens
Metabolic Regulation in Response to Pests and Pathogens
When pests and pathogens threaten plant health, nature's biochemical defenses come to the forefront. Plants employ a range of metabolic strategies to fend off these adversaries. Our sophisticated instrumentation, such as the Thermo Scientific Q Exactive Plus LC-MS/MS and the Agilent 7890B GC-MS, provides a window into this biochemical battlefield. Here, we explore the world of defensive compounds, including alkaloids, terpenoids, and phenolic compounds, and understand how plants adapt their metabolic pathways in response to biotic stressors.
- Identification of Defense Compounds: Metabolomics identifies the specific secondary metabolites produced by plants when under attack by pests or pathogens. This includes alkaloids, terpenoids, flavonoids, and other compounds known for their defensive properties.
- Quantification of Metabolite Levels: We quantify changes in metabolite levels during biotic stress responses. This quantification reveals the magnitude of metabolic changes, providing insights into the severity of the stress and the effectiveness of plant defenses.
- Pathway Analysis: Metabolomics allows us to trace the metabolic pathways involved in plant defense responses. This includes understanding how plants reroute metabolic resources to bolster defenses, often at the expense of growth and reproduction.
 Metabolic Crosstalk in Plant-Mycorrhizal Fungi Symbiosis
Metabolic Crosstalk in Plant-Mycorrhizal Fungi Symbiosis
Plant-mycorrhizal fungi symbiosis exemplifies the intricate dance of cooperation in nature. Plants provide mycorrhizal fungi with carbon compounds, while fungi enhance the plant's nutrient uptake, especially phosphorus. To unravel the metabolic intricacies of this mutually beneficial relationship, we turn to advanced instrumentation, including the Waters Xevo G2-XS QTof LC-MS and the Shimadzu GCMS-TQ8050.
- Nutrient Transfer: Metabolomics elucidates the metabolites involved in nutrient transfer between plants and mycorrhizal fungi. Specifically, it reveals how plants provide sugars and lipids to fungi and how fungi facilitate the uptake of nutrients, such as phosphorus and nitrogen, by plants.
- Stress Tolerance: We analyze the metabolic changes in both plants and mycorrhizal fungi in response to environmental stressors, such as drought or nutrient deficiency. This helps us understand how symbiosis enhances stress tolerance.
- Overall Plant Health: Metabolomics provides insights into the overall health of plants engaged in symbiosis. By profiling metabolites, we can assess the impact of the symbiotic relationship on plant metabolism and vitality.
Metabolomic Techniques for Plant-Environment Interaction Studies
Liquid Chromatography-Mass Spectrometry (LC-MS): LC-MS enables the comprehensive profiling of metabolites in plant samples. It aids in the detection and quantification of compounds, including secondary metabolites, nutrients, and harmful substances, offering insights into how plants adapt metabolically to changing environmental conditions.
- Instrument Models: Prominent LC-MS instruments include the Agilent 1290 Infinity II LC-MS and the Thermo Scientific Q Exactive Plus LC-MS/MS.
Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS specializes in the analysis of volatile compounds, making it valuable for studying plant responses to specific environmental factors. It is particularly useful for investigating the release of volatile organic compounds by plants under stress.
- Instrument Models: Widely used GC-MS models encompass the Agilent 7890B GC-MS and the Shimadzu GCMS-TQ8050.
Case. Metabolomics Analysis of Portuguese Common Bean Accessions Grown in Contrasting Environments
Background
This study aimed to investigate the metabolomics diversity of Portuguese common bean accessions grown in two different environments and identify specific metabolites that could be associated with heat tolerance in these beans.
Samples
The study involved 107 underexploited Portuguese common bean accessions, which were cultivated in two contrasting environments: Cabrela (traditional) and Cordoba (heat-stress).
Technological Methods
Metabolite Identification: Metabolites were identified primarily using negative ionization mode, which offers improved sensitivity and lower detection limits. Online databases were used for annotation, and compounds were selected based on VIP scores (Variable Importance in Projection) higher than 0.8.
Annotation Challenges: The annotation of compounds was hindered by limited diversity in available online libraries and the quality of mass spectrometry (MS) and MS/MS fragmentation spectra. This was due to the historical underrepresentation of legume metabolomics research.
Superclasses Classification: The annotated compounds were categorized into seven different superclasses, including phenylpropanoids and polyketides, organic oxygen compounds, benzenoids, lipids, nucleosides, nucleotides, organic acids, and derivatives.
Results:
- Metabolic Diversity: Despite the absence of significant differences in total phenolic and flavonoid content between common bean accessions grown in different environments, individual metabolites, such as benzenoids, lipids, and organoheterocyclic compounds, exhibited significant variations.
- Impact of Morphological Traits: Accessions with colored seeds displayed higher metabolite diversity compared to white-seeded accessions, both in Cabrela and Cordoba. Within each environment, two distinct clusters were identified based on metabolite abundance.
- Gene Pool of Origin: Accessions with Mesoamerican origins generally showed lower metabolite content than those with Andean or mixed origins, highlighting the potential of Portuguese common bean germplasm for future breeding programs.
- Breeding Implications: Accessions rich in specific metabolites influenced by accession effects, such as astragalin and quercetin, may be valuable for breeding programs aimed at developing varieties with enhanced nutraceutical properties. Additionally, accessions with metabolites highly influenced by environmental conditions, like salicylic acid, could be targeted for breeding in heat-stress environments.
- Metabolite Interactions: Complex interactions were observed between metabolites belonging to different superclasses, shedding light on shared metabolic pathways and potential metabolic regulation in common beans.
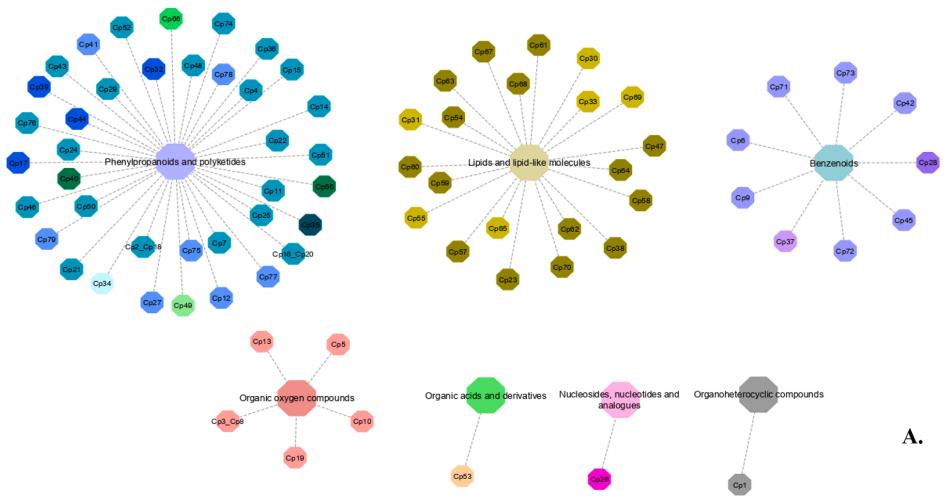 Classification of metabolites into different superclasses.
Classification of metabolites into different superclasses.
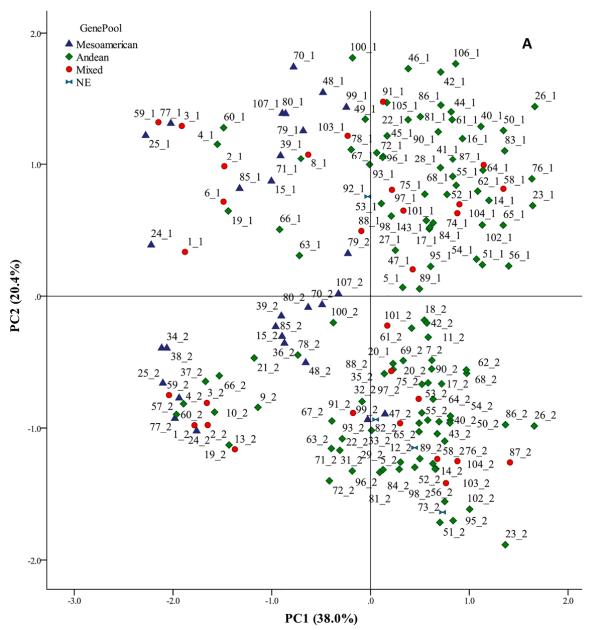 Score plot obtained by principal component analysis (PCA), showing common bean accessions, cropped in contrasting environments.
Score plot obtained by principal component analysis (PCA), showing common bean accessions, cropped in contrasting environments.
Reference
- Mecha, Elsa, et al. "Metabolomics profile responses to changing environments in a common bean (Phaseolus vulgaris L.) germplasm collection." Food Chemistry 370 (2022): 131003.


 Classification of metabolites into different superclasses.
Classification of metabolites into different superclasses. Score plot obtained by principal component analysis (PCA), showing common bean accessions, cropped in contrasting environments.
Score plot obtained by principal component analysis (PCA), showing common bean accessions, cropped in contrasting environments.

