Nucleotide metabolism encompasses a series of interconnected biochemical reactions responsible for the synthesis, degradation, and regulation of nucleotides - the building blocks of nucleic acids (DNA and RNA). This intricate network of processes plays a pivotal role in fundamental biological functions, including DNA replication, RNA transcription, protein synthesis, and cellular signaling. Disturbances in nucleotide metabolism can lead to severe health issues, making its analysis crucial for understanding cellular health and disease mechanisms.
Nucleotide metabolism analysis holds immense significance in advancing our understanding of plant biology. By unraveling the intricate pathways involved in nucleotide synthesis, salvage, and degradation, researchers can decipher the underlying mechanisms that influence plant responses to stressors, nutrient availability, and disease. Additionally, studying nucleotide metabolism aids in identifying potential targets for crop improvement, biotechnology applications, and sustainable agriculture practices.
Creative Proteomics' Plant Nucleotide Metabolism Analysis Projects
Nucleotide Biosynthesis Pathway Analysis: Unravel the steps of nucleotide creation in plants, identifying key components that drive growth and development.
Nucleotide Salvage Pathway Analysis: Investigate how plants recycle nucleotides, optimizing resources and enhancing resilience against stressors.
Nucleotide Degradation Pathway Analysis: Study the breakdown of nucleotides, revealing insights into nutrient recycling and energy management.
Nucleotide Transport and Compartmentalization Analysis: Explore nucleotide movement within plant cells, uncovering vital regulatory processes.
Nucleotide Metabolism in Stress Responses Analysis: Examine nucleotide shifts during stress, aiding the understanding of plant adaptability and survival mechanisms.
Nucleotide Metabolism in Developmental Processes Analysis: Discover how nucleotides influence growth stages, from germination to reproduction.
Nucleotide Signaling and Communication Analysis: Probe nucleotides as signals, illuminating their role in coordinating plant physiology and responses.
Comparative Nucleotide Metabolism Analysis: Compare nucleotide pathways across species, revealing evolutionary insights and potential for trait improvement.
Quantitative and Qualitative Data Integration: Creative Proteomics employs robust methodologies to combine quantitative measurements of nucleotide concentrations with qualitative information about metabolic intermediates and pathway dynamics. This integrative approach provides a holistic view of nucleotide metabolism, enriching our understanding of plant cellular processes.
Nucleotide Metabolism Analysis Techniques
Mass Spectrometry-Based Profiling
Utilizing state-of-the-art mass spectrometers, including the Agilent 6545 Q-TOF LC/MS and the Thermo Fisher Q Exactive HF-X Hybrid Quadrupole-Orbitrap Mass Spectrometer, Creative Proteomics offers high-resolution profiling of nucleotide metabolites. This approach enables the precise quantification and qualitative characterization of a wide range of nucleotide species, facilitating the identification of metabolic shifts and regulatory mechanisms.
Isotope Labeling and Tracing
Isotope labeling and tracing techniques provide a dynamic view of nucleotide metabolism. By introducing stable isotopes into plant samples, Creative Proteomics can trace the fate of nucleotide precursors and intermediates. This technique allows researchers to discern metabolic fluxes, pathways, and reaction rates, shedding light on the kinetics of nucleotide synthesis and turnover.
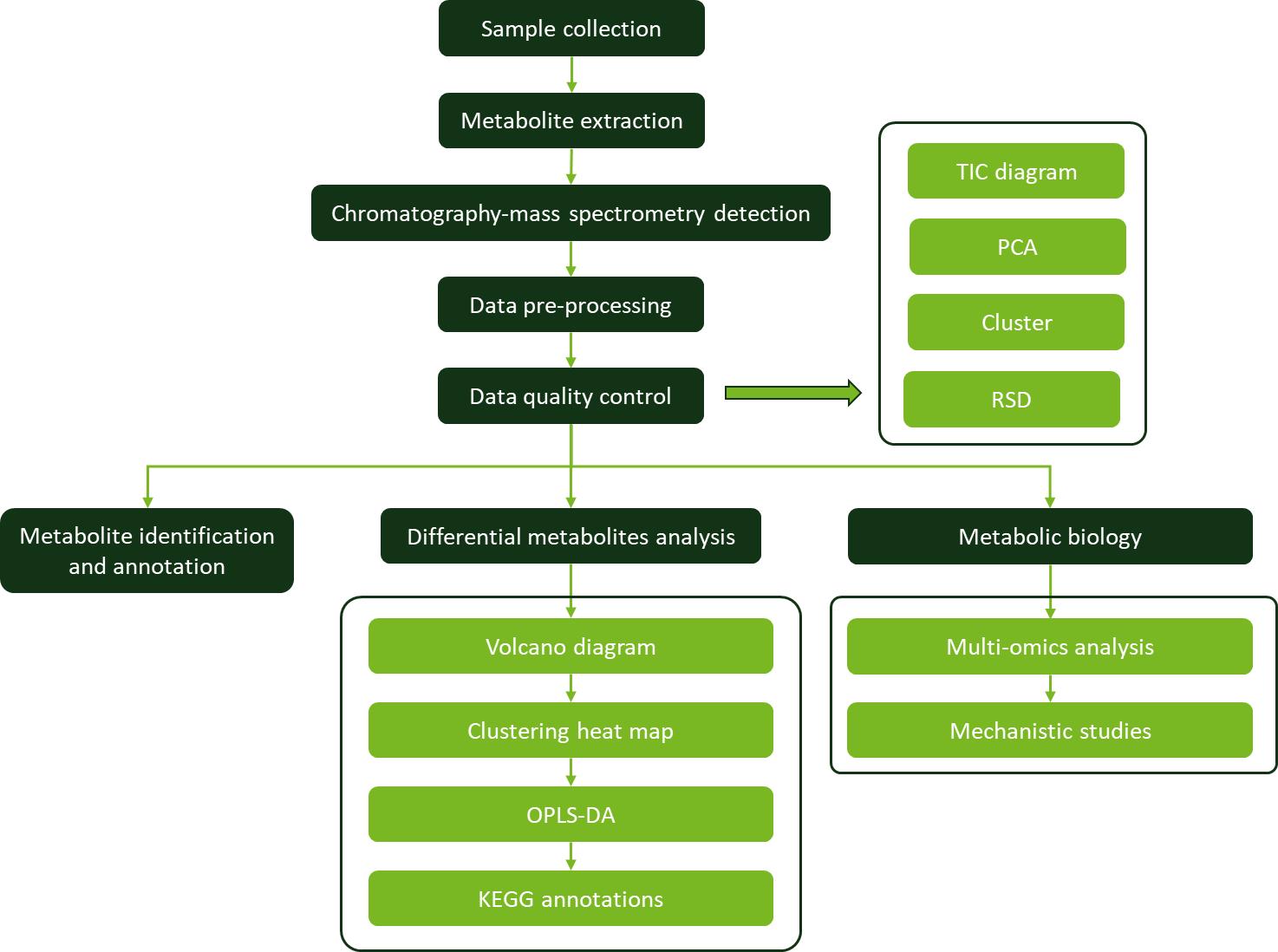 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
List of Nucleotide Analyzed (including but not limited to)
Purine Nucleotides
| Nucleotide Class |
Representative Metabolites |
| Adenylates (AMP, ADP, ATP) |
Adenosine monophosphate (AMP), Adenosine diphosphate (ADP), Adenosine triphosphate (ATP) |
| Guanylates (GMP, GDP, GTP) |
Guanosine monophosphate (GMP), Guanosine diphosphate (GDP), Guanosine triphosphate (GTP) |
| Inosinates (IMP, IDP, ITP) |
Inosine monophosphate (IMP), Inosine diphosphate (IDP), Inosine triphosphate (ITP) |
Pyrimidine Nucleotides
| Nucleotide Class |
Representative Metabolites |
| Cytidylates (CMP, CDP, CTP) |
Cytidine monophosphate (CMP), Cytidine diphosphate (CDP), Cytidine triphosphate (CTP) |
| Uridylates (UMP, UDP, UTP) |
Uridine monophosphate (UMP), Uridine diphosphate (UDP), Uridine triphosphate (UTP) |
| Thymidylates (TMP, TDP, TTP) |
Thymidine monophosphate (TMP), Thymidine diphosphate (TDP), Thymidine triphosphate (TTP) |
Other Nucleotide Metabolites
| Nucleotide Class |
Representative Metabolites |
| Xanthosine Nucleotides |
Xanthosine monophosphate (XMP) |
| Deoxyadenylates (dAMP, dADP, dATP) |
Deoxyadenosine monophosphate (dAMP), Deoxyadenosine diphosphate (dADP), Deoxyadenosine triphosphate (dATP) |
| Deoxyguanylates (dGMP, dGDP, dGTP) |
Deoxyguanosine monophosphate (dGMP), Deoxyguanosine diphosphate (dGDP), Deoxyguanosine triphosphate (dGTP) |
| Deoxycytidylates (dCMP, dCDP, dCTP) |
Deoxycytidine monophosphate (dCMP), Deoxycytidine diphosphate (dCDP), Deoxycytidine triphosphate (dCTP) |
| Deoxyuridylates (dUMP, dUDP, dUTP) |
Deoxyuridine monophosphate (dUMP), Deoxyuridine diphosphate (dUDP), Deoxyuridine triphosphate (dUTP) |
Application of Our Nucleotide Analysis Service
Crop Improvement: Understanding nucleotide metabolism aids in developing crops with enhanced stress tolerance, yield, and nutritional content.
Plant-Microbe Interactions: Analyzing nucleotide metabolism sheds light on the plant's response to microbial pathogens and symbiotic associations.
Environmental Adaptation: Studying nucleotide metabolism provides insights into how plants adapt to changing environmental conditions, such as nutrient availability and climate fluctuations.
Sample Requirements for Nucleotide Assay
| Sample Types |
Minimum Sample Size |
| Plant Samples |
Roots, stems and leaves, floral parts, fruits/seeds, rhizomes, buds/tender leaves, tissue sections, pollen, bark, trunk/wood, resin/gum, resin acids, seedlings/young plants, rhizosphere soil, root exudates. |
50 mg - 1 g |
| Animal Samples |
Serum/Plasma |
200-500 μL |
| Tissues |
20-100 mg |
| Urine/Feces |
1-2 mL / 100 mg |
| Cell Samples |
Cells and Culture |
106 - 108 cells |
Case 1. Unlocking Plant Metabolism: Comprehensive Analysis of Nucleotides and Nucleosides using Mass Spectrometry
Background:
Mass spectrometry-based metabolomics is a powerful approach utilized to study the composition and dynamics of metabolites, including nucleotides (NTs) and nucleosides (Ns), in complex biological samples. This technique allows for the sensitive detection, identification, and quantification of these important molecules, providing insights into their roles in plant biology and metabolism.
Samples:
The samples analyzed in mass spectrometry-based metabolomics studies include plant tissues and extracts. These samples are subjected to various sample preparation techniques, such as liquid-liquid extraction (LLE) and solid-phase extraction (SPE), to isolate and enrich NTs and Ns. The choice of sample preparation method depends on factors like the nature of the analytes and the complexity of the matrix.
Methods:
Sample Extraction and Preparation:
Plant samples, typically leaves or other tissues, undergo a meticulous extraction process to access NTs and Ns. Initially, the tissues are homogenized to create a representative sample. Subsequently, metabolite extraction is carried out using appropriate solvents to ensure efficient release of the target compounds. Two prevalent techniques, liquid-liquid extraction (LLE) and solid-phase extraction (SPE), are commonly employed. SPE offers distinct advantages by selectively binding and concentrating the analytes while eliminating unwanted matrix components. This step ensures reduced interference and enhanced sensitivity during downstream analysis.
Chromatographic Separation:
Chromatography serves as a crucial step in isolating NTs and Ns from the complex mixture of metabolites. Standard reverse-phase chromatography, employing C8 or C18 columns, is a common approach. These hydrophobic columns are often modified with polar groups to facilitate the retention of polar metabolites, including NTs. An alternative technique, hydrophilic interaction liquid chromatography (HILIC), excels in separating polar and charged metabolites. Porous graphitized carbon (PGC) chromatography provides high-resolution separation for various polar compounds, such as NTs. The choice of chromatographic method depends on the chemical properties of the analytes and the specific research objectives.
Mass Spectrometry Detection and Analysis:
Mass spectrometry is the cornerstone of NT and N analysis, with instruments like triple-quadrupole (QqQ) and high-resolution mass spectrometers (HRMS) being commonly employed. The selection of ionization mode, whether positive or negative, hinges on factors such as analyte charge and matrix effects. The negative mode is often preferred for its informative fragmentation patterns. To enhance ionization efficiency, additives like hexafluoroisopropanol and methanol are incorporated as charge carriers. However, adduct formation and in-source decay can influence sensitivity and signal accuracy, necessitating thoughtful optimization of source conditions.
Tandem Mass Spectrometry and Fragmentation:
Tandem mass spectrometry (MS/MS) is employed for its ability to provide structural insights into NTs and Ns. This process involves selecting a precursor ion for fragmentation within the collision cell. Notably, NTs and Ns exhibit distinct fragmentation pathways that allow for the prediction of product ions. Single reaction monitoring (SRM) is utilized to quantify specific transitions, and the collision energy voltage is meticulously adjusted to ensure accurate quantification. The development of fragmentation patterns and the use of databases aid in the identification and quantification of target metabolites.
Data Analysis and Interpretation:
Data analysis encompasses several steps, starting with peak detection based on retention time and mass-to-charge ratio (m/z). The identification of metabolites relies on spectral matching against databases and the comparison to standard compounds. Quantification is achieved through the use of internal standards and calibration curves. Pathway analysis contextualizes the results, shedding light on metabolic networks and interactions involving NTs and Ns. Advanced software tools streamline data processing and visualization, facilitating robust interpretation.
Instrument Performance and Sensitivity:
Triple-quadrupole instruments excel in sensitivity and are commonly employed for targeted quantification of NTs and Ns. High-resolution mass spectrometers offer precise measurements, making them invaluable for non-targeted analyses. Sensitivity is influenced by factors such as matrix effects, ion load, and source conditions. Careful optimization of sample preparation, chromatography, and instrument parameters is pivotal in maximizing sensitivity and ensuring accurate quantification.
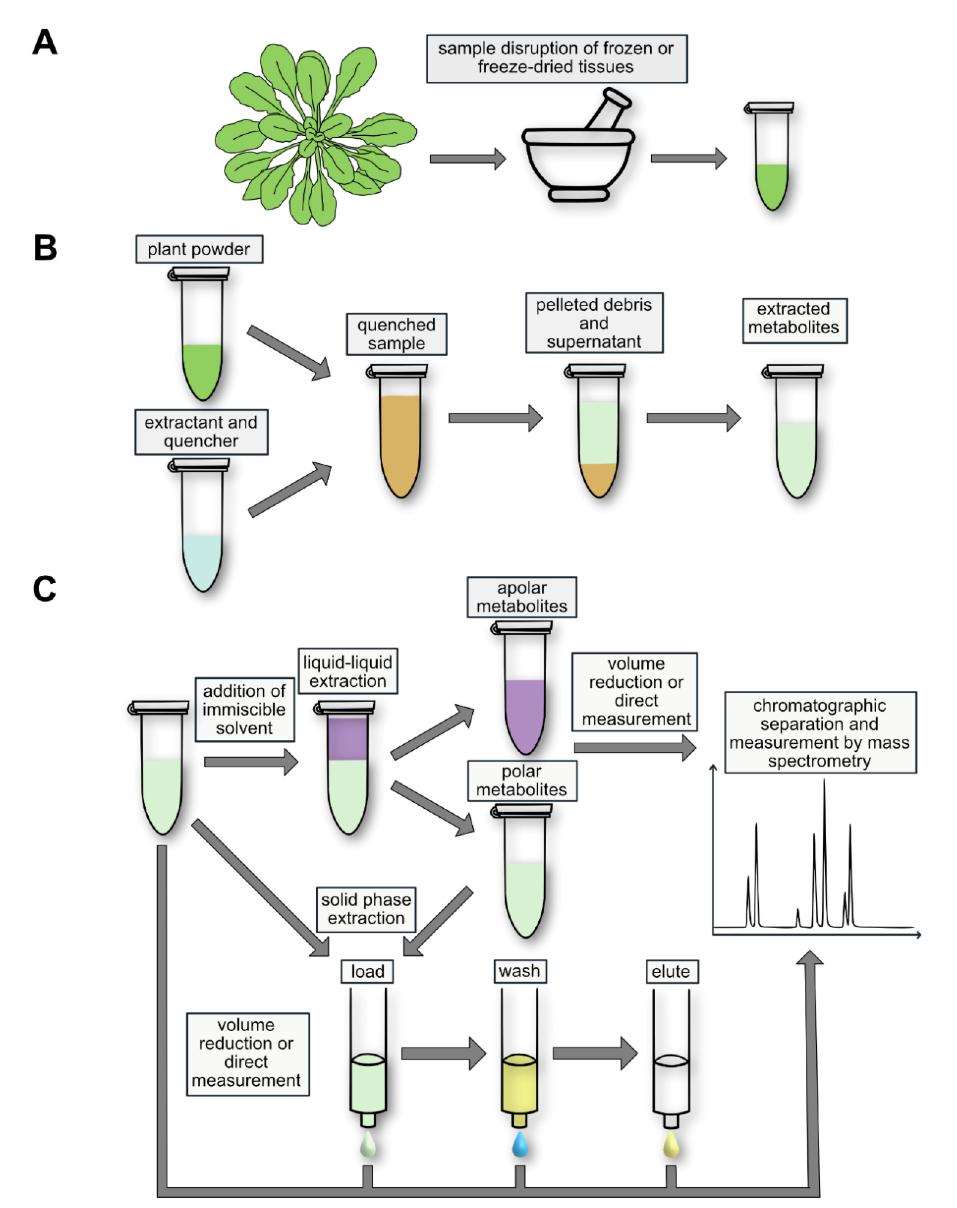 Schematic representation of a typical workflow for the analysis of nucleotides and nucleosides in plants
Schematic representation of a typical workflow for the analysis of nucleotides and nucleosides in plants
Results
Mass spectrometry-based metabolomics provides detailed insights into the presence and levels of NTs and Ns in plant samples. The technique allows for the identification and quantification of a wide range of NTs and Ns, both native and modified forms. Data analysis reveals the variations in metabolite profiles, enabling the elucidation of metabolic pathways and networks involving NTs and Ns. Challenges in sensitivity, matrix effects, and fragmentation are being addressed, and ongoing developments in instrumentation and data analysis are expected to further advance our understanding of the roles of NTs and Ns in plant physiology.
Reference
- Straube, Henryk, Claus-Peter Witte, and Marco Herde. "Analysis of Nucleosides and Nucleotides in Plants: An Update on Sample Preparation and LC–MS Techniques." Cells 10.3 (2021): 689.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service Schematic representation of a typical workflow for the analysis of nucleotides and nucleosides in plants
Schematic representation of a typical workflow for the analysis of nucleotides and nucleosides in plants

