Exploring the World of Quinones and Their Metabolites
In the realm of organic chemistry, quinones stand as intriguing compounds that captivate researchers and enthusiasts alike. Characterized by their unique cyclic structure featuring a double bond and two carbonyl groups, quinones play multifaceted roles in the intricate tapestry of biological processes. These compounds are not merely confined to chemical reactions; they hold key positions in electron transfer, enzymatic orchestration, and the delicate equilibrium of oxidative stress regulation. Furthermore, quinones emerge as pivotal players in both endogenous and exogenous compound metabolism, influencing fundamental pathways that impact our health and well-being.
Understanding quinone metabolism is crucial for unraveling their implications in health and disease. Quinones are associated with oxidative damage, cancer, neurodegenerative disorders, and cardiovascular diseases. Analyzing quinones and their metabolites can shed light on potential biomarkers, therapeutic targets, and mechanistic pathways. In addition, quinones act as active participants in oxidative processes, influencing plant growth, defense mechanisms, and adaptive strategies. An in-depth study of quinone analysis will not only unravel the complexity of plant biochemistry, but also has the potential to uncover clues about stress responses, metabolic pathways, and physiological adaptations in the plant world.
Quinones Detection Projects at Creative Proteomics
Creative Proteomics presents an array of quinones analysis services that transcend the conventional boundaries of inquiry. Our mission is to unravel the mysteries surrounding quinones and their metabolites, shedding light on their roles within the intricate web of cellular dynamics. Our suite of services encompasses a comprehensive spectrum of quinones analysis, including:
Quinone Identification: Armed with cutting-edge mass spectrometry technologies, Creative Proteomics embarks on precise identification journeys, unraveling the enigmatic identities of quinones and their elusive metabolites lurking within complex biological samples.
Quantitative Profiling: Harnessing advanced quantification methodologies, we navigate the quantitative landscape of quinones, elucidating their levels and concentrations with an unwavering commitment to accuracy.
Metabolic Pathway Mapping: Our scientific explorations extend beyond individual compounds, encompassing the broader canvas of metabolic pathways. Creative Proteomics is dedicated to unraveling the interconnectedness of quinones with other biomolecules, painting a holistic portrait of cellular orchestration.
Technologies Employed for Quinones Analysis
Liquid Chromatography-Mass Spectrometry (LC-MS): Creative Proteomics employs LC-MS techniques for high-resolution separation and accurate mass determination of quinones and their metabolites. Instruments such as the Thermo Scientific Q Exactive series are used for precise analysis.
Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS is utilized for volatile quinone metabolite analysis, providing detailed structural information. Instruments like the Agilent 7890A GC system coupled with mass spectrometers ensure robust quantification.
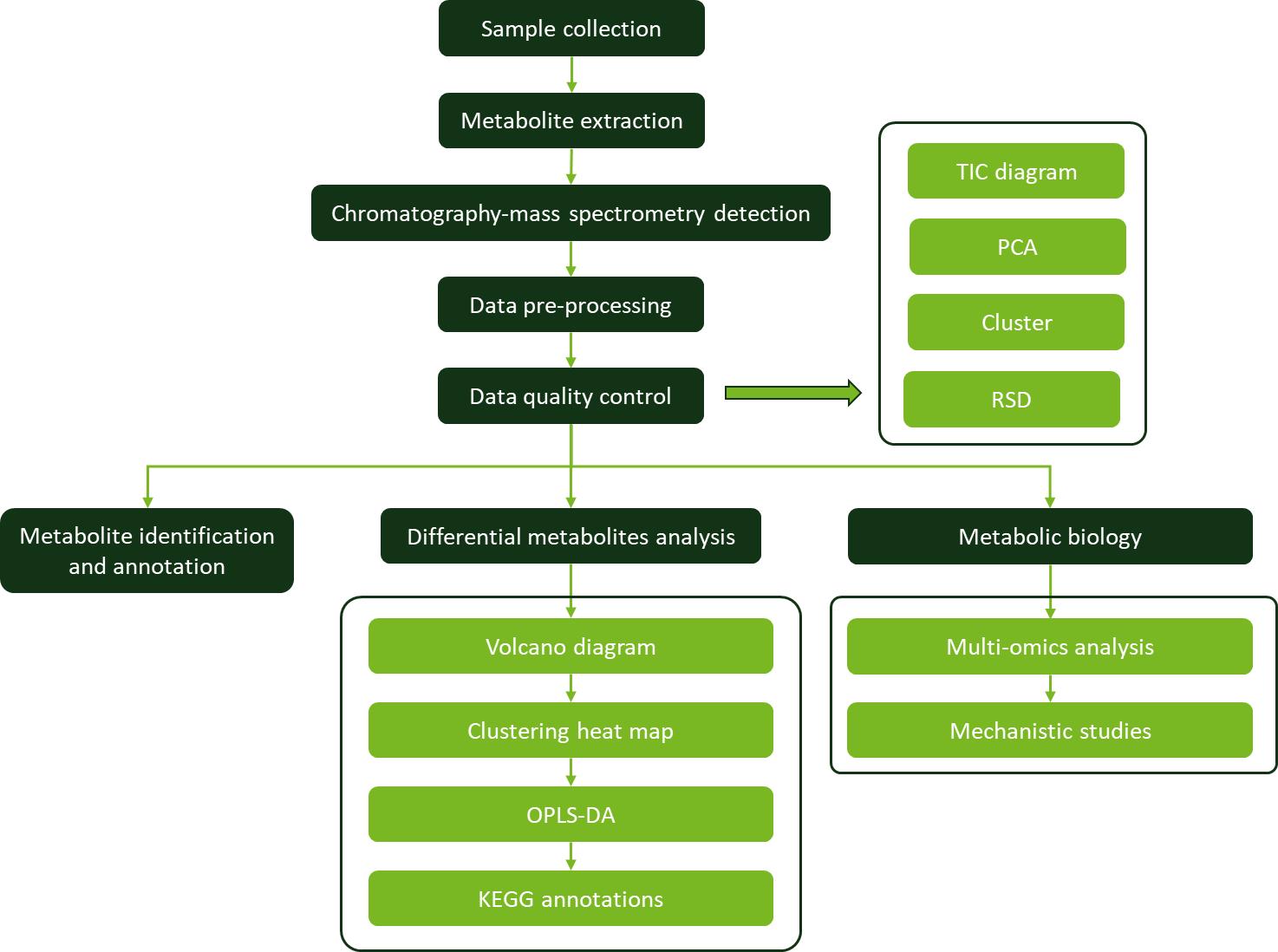 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
List of Quinones Analyzed (including but not limited to)
| Types |
Quinone Compounds |
| Naphthoquinones |
Plumbagin, Juglone, Lapachol, Menadione, Lawsone, Alizarin |
| Benzoquinones |
Hydroquinone, Coenzyme Q1, Coenzyme Q2, Coenzyme Q3, Coenzyme Q4, Coenzyme Q5 |
| Ubiquinones |
Ubiquinone-1 (UQ-1), Ubiquinone-2 (UQ-2), Ubiquinone-3 (UQ-3), Ubiquinone-4 (UQ-4), Ubiquinone-5 (UQ-5), Ubiquinone-6 (UQ-6) |
| Plastoquinones |
Plastoquinone-1, Plastoquinone-2, Plastoquinone-3, Plastoquinone-4, Plastoquinone-5, Plastoquinone-6 |
| Other Quinones |
Emodin, Menadione Sodium Bisulfite, Embelin, Toluquinol |
Sample Requirements for Quinones Assay
| Sample Types |
Minimum Sample Size |
| Plant Samples |
Roots, stems and leaves, floral parts, fruits/seeds, rhizomes, buds/tender leaves, tissue sections, pollen, bark, trunk/wood, resin/gum, resin acids, seedlings/young plants, rhizosphere soil, root exudates. |
50 mg - 1 g |
| Animal Samples |
Urine |
1-2 mL |
| Serum/Plasma |
100-500 µL |
Applications of Quinones Metabolomics Analysis
Plant Biochemistry and Agriculture: In plant biochemistry and agriculture, quinones play an integral role in secondary metabolism, actively contributing to processes such as plant defense mechanisms, UV protection, and chemosensitization. In-depth study of different quinone profiles through metabolomics analysis deepens our understanding of plant responses to stressors.
Environmental Science and Toxicology: Quinones are important biomarkers of environmental pollution, and meticulous metabolomic analysis allows for meticulous tracking of quinone fluctuations in response to environmental changes, contributing to accurate environmental monitoring and comprehensive risk assessment. In addition, the key role of quinones in oxidative stress pathways enriches toxicological studies and paves the way for the development of complex pollution mitigation strategies.
Human Health and Disease Research: Quinones have been linked to a wide range of diseases, from cancer to cardiovascular disease, and metabolomic analyses have revealed biomarkers associated with quinones. In addition, the pharmaceutical potential of quinones, particularly in anticancer therapy, has been boosted by the identification of their unique properties. Metabolomics approaches can accelerate drug discovery and development.
Microbial ecology and biotechnology: Quinones play an integral role in microbial redox reactions, energy transfer and bioproduct synthesis. Uncovering quinone-mediated metabolic pathways in microbes to optimize microbial production and usher in a new era of eco-friendly innovation.
Case 1. Enhanced Secondary Metabolite Production and Anti-Inflammatory Activity in Aloe vera Adventitious Roots through Salicylic Acid Elicitation
Background:
The study aimed to investigate the metabolic changes and anti-inflammatory activity enhancement in Aloe vera adventitious roots through SA (salicylic acid) elicitation. Previous attempts at Aloe vera cell culture were limited due to phenolic compound-related cell death, necessitating the development of an optimized suspension culture system.
Samples:
Adventitious roots of Aloe vera were used as the experimental material. The study involved optimizing the suspension culture conditions for these roots and examining the effects of SA, MJ (methyl jasmonate), and ethephon on the production of secondary metabolites, particularly aloe emodin and chrysophanol.
Methods:
1. Optimization of Suspension Culture:
Three-week-old adventitious roots from Aloe vera were transferred to liquid MS media with 30 g/L sucrose and supplemented with different plant growth regulators (0.5 mg/L IBA, IAA, or NAA).
After four weeks of culture, the growth and performance of the adventitious roots were assessed in different media compositions (MS, 1/2 MS, 2MS, B5, SH).
2. Elicitation and Metabolite Analysis:
SA, MJ, and ethephon were added to 35-day-old adventitious roots at various concentrations.
Production of aloe emodin and chrysophanol was quantified in both the adventitious roots and the growth medium over a specific time course.
Metabolite analysis was performed using UPLC-ESI/MS/MS to identify compounds induced by SA elicitation. Data analysis included PCA, PLS-DA, and OPLS-DA to assess metabolite alterations.
GC-MS and LC-MS were used to analyze primary metabolites, focusing on the tricarboxylic acid cycle (TCA) and malonyl-CoA, a precursor for tricyclic aromatic quinone biosynthesis.
3. Gene Expression Analysis:
Full-length cDNAs for Aloe vera OKS (AvOKS) and OKS like-1 (AvOKSL-1) were isolated from adventitious roots based on sequences from Aloe arborescense OKS (AaOKS) and PKS4 (AaPKS4).
Real-time PCR and RT-qPCR were employed to analyze the expression levels of AvOKS and AvOKSL-1 in response to SA treatment.
4. Anti-Inflammatory Activity Assay:
Extracts from adventitious roots treated with different concentrations of SA were tested for anti-inflammatory activity using JB6 P+ cells and a UVB-induced skin cell model.
Promoter activity of COX-2, as well as transactivation of NF-κB and AP-1, was measured to assess the anti-inflammatory effects of the extracts.
5. Data Analysis:
Various statistical analyses, such as ANOVA, were applied to compare metabolite levels, gene expression, and anti-inflammatory effects between different treatments.
Multivariate analysis, including PCA, PLS-DA, and OPLS-DA, was conducted to identify differences and patterns in metabolite profiles induced by SA treatment.
6. Compound Identification:
UPLC-ESI/MS/MS was employed to obtain structural information on the SA-induced compounds. MS/MS fragmentation patterns were analyzed to tentatively identify compounds, including chrysophanol and aloe emodin.
Results
SA Elicitation: SA elicitation significantly increased the production of aloe emodin and chrysophanol, with up to 10-11 fold and 5-13 fold increases, respectively, within 24 hours.
Metabolite Identification: UPLC-ESI/MS/MS identified 37 SA-induced compounds, including potential tricyclic aromatic quinone derivatives.
Gene Expression and Malonyl-CoA: Gene expression of AvOKS and AvOKSL-1 was upregulated by SA treatment, correlating with increased malonyl-CoA content and suggesting their involvement in tricyclic aromatic quinone biosynthesis.
Enhanced Anti-Inflammatory Activity: Extracts from SA-treated adventitious roots showed enhanced anti-inflammatory activity in UVB-treated mouse skin cells, possibly attributed to SA-induced metabolites.
In summary, the study established an optimized suspension culture system for Aloe vera adventitious roots, demonstrated the significant impact of SA elicitation on secondary metabolite production, identified SA-induced compounds through advanced metabolomics techniques, and showed the potential of SA-induced metabolites for enhancing anti-inflammatory activity.
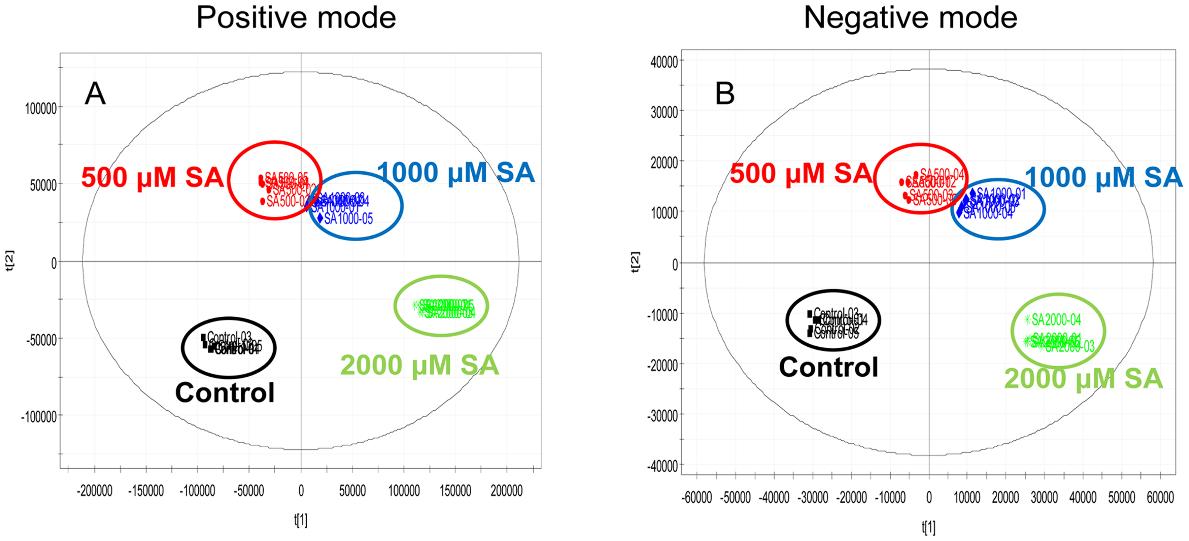 OPLS-DA score plot.
OPLS-DA score plot.
Reference
- Lee, Yun Sun, et al. "Enhancement of anti-inflammatory activity of Aloe vera adventitious root extracts through the alteration of primary and secondary metabolites via salicylic acid elicitation." PloS one 8.12 (2013): e82479.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service OPLS-DA score plot.
OPLS-DA score plot.

