Plant metabolomics is a branch of science that focuses on the comprehensive analysis of small molecule metabolites present within plants. Metabolites play a crucial role in mediating various physiological and biochemical processes within plants, including nutrient uptake, stress response, energy production, and secondary metabolite synthesis. By studying the diverse array of metabolites present in a plant, researchers can gain valuable insights into its overall health, development, and interactions with its environment.
At Creative Proteomics, we recognize the immense potential of plant metabolomics analysis in unraveling the complexities of plant biology. Our services aim to bridge the gap between metabolite profiling and functional interpretation, enabling researchers to make informed decisions and advancements in agriculture, biotechnology, and environmental science.
Plants Metabolomics Projects by Creative Proteomics
Our expertise is finely tuned to address the complexities of plant metabolomics, offering tailored services that empower researchers and industries to delve deeper into the biochemical intricacies of plant life.
Stress-Responsive Metabolite Profiling
Stress is an inevitable challenge that plants face in their natural environment. Creative Proteomics specializes in conducting metabolomic analyses to unveil the intricate changes that occur in plant metabolites under various stress conditions. Whether it's drought, salinity, or pathogen attack, our advanced techniques reveal the dynamic responses of plants at the molecular level. By identifying stress-specific metabolites, we provide crucial insights for developing strategies to enhance plant resilience and promote sustainable agriculture.
Secondary Metabolite Characterization
Secondary metabolites are the essence of a plant's unique chemical fingerprint. Creative Proteomics excels in the comprehensive characterization of these compounds, including flavonoids, alkaloids, and terpenoids. Our state-of-the-art mass spectrometry-based analysis allows us to precisely identify and quantify secondary metabolites, shedding light on their roles in plant defense, pigmentation, and signaling. Through this characterization, we contribute to unraveling the intricate web of plant adaptation and interactions.
Nutrient Metabolism Investigations
Efficient nutrient utilization is fundamental to optimal plant growth and productivity. Creative Proteomics offers in-depth analyses of nutrient metabolism, uncovering the underlying pathways that govern nutrient uptake, assimilation, and utilization. By deciphering the metabolic intricacies involved in nutrient management, we provide valuable insights that pave the way for enhancing crop yields, nutrient efficiency, and sustainable agricultural practices.
Comparative Metabolomics Studies
Bridging the gaps between species and varieties, Creative Proteomics conducts comparative metabolomics studies that offer a broader perspective on plant metabolic traits. Our comprehensive analyses reveal unique metabolic signatures, evolutionary adaptations, and metabolic diversity across different plant groups. Through these cross-species investigations, we contribute to the understanding of plant evolution, genetic diversity, and the potential for crop improvement.
Hormone-Regulated Metabolite Profiling
Plant hormones orchestrate a myriad of physiological processes, from growth and development to responses to external stimuli. Creative Proteomics specializes in studying the metabolomic changes induced by plant hormones, such as auxins, gibberellins, and abscisic acid. By dissecting the intricate interplay between hormones and metabolites, we provide insights into hormone signaling pathways and their roles in shaping plant phenotypes.
Which Plant Metabolomics Analysis Method to Choose?
| Aspect |
Plant Targeted Metabolomics |
Plant Untargeted Metabolomics |
| Research Objective |
Study specific metabolites or pathways of interest |
Explore entire metabolite profile |
| Analytical Focus |
Known metabolites |
Broad range of metabolites |
| Biomarker Discovery |
Identifies and validates specific biomarkers |
May uncover potential biomarkers |
| Translational Research |
Studies metabolic pathway changes and regulation |
Reveals diverse metabolic responses under various conditions |
| Data Interpretation |
Emphasizes known metabolite identification and quantification |
Complex data analysis, discovering patterns |
| Applications |
Pathway analysis, focused studies |
Biomarker discovery, holistic insights |
| Advantages |
Accurate quantification, focused research |
Discovery of novel metabolites, broad insights |
| Practical Considerations |
Well-defined research questions |
Exploratory research |
| Decision Considerations |
Specific goals, research budget |
Scope of research, data analysis capacity |
| Combined Approach |
Possible for in-depth confirmation after untargeted profiling |
Followed by targeted analysis for validation |
| Follow-Up Studies |
Mechanistic investigations, targeted studies |
Provides insights for broader studies, further exploration |
Plants Metabolomics Analysis Techniques
Central to the success of plants metabolomics analysis are state-of-the-art mass spectrometry techniques. Creative Proteomics employs cutting-edge instruments to unravel the intricate world of plant metabolites.
- Liquid Chromatography-Mass Spectrometry (LC-MS): LC-MS combines the separation capabilities of liquid chromatography with the sensitivity and specificity of mass spectrometry. Instruments such as Thermo Scientific Q Exactive™ HF-X enable high-resolution and accurate mass measurements.
- Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS is ideal for volatile and semi-volatile compound analysis. The Agilent 7890B GC System coupled with mass spectrometry provides unparalleled sensitivity and reproducibility.
- Nuclear Magnetic Resonance (NMR) Spectroscopy: NMR spectroscopy offers insights into the structural elucidation of metabolites. Our Bruker Avance III HD 600 MHz NMR Spectrometer ensures precise and detailed analysis.
- High-Resolution Mass Spectrometry: Instruments like Thermo Scientific Orbitrap Exploris 480 provide exceptional mass accuracy and resolution, facilitating the detection of a wide range of metabolites.
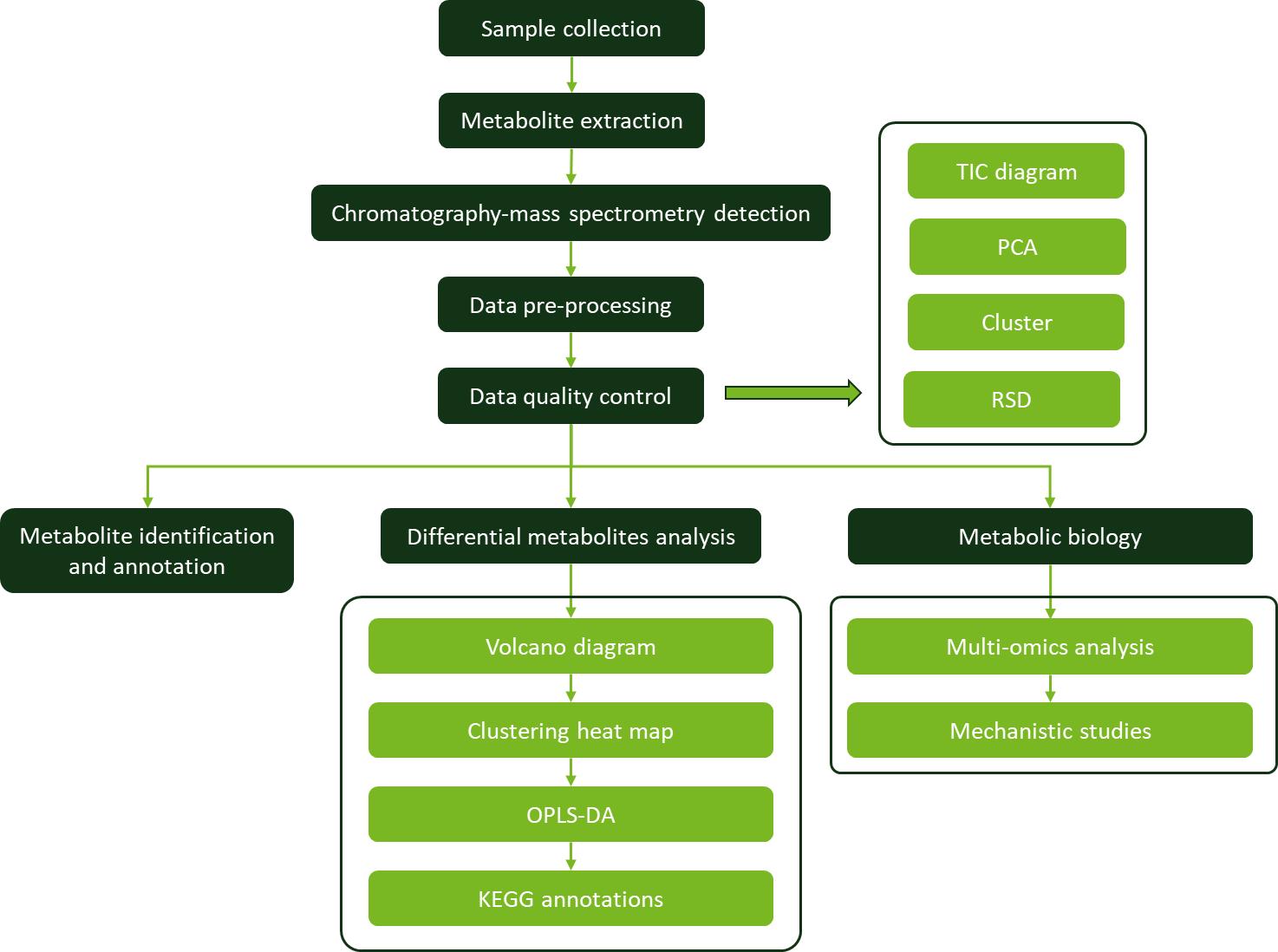 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Applications of Plants Metabolomics Analysis
Plant Physiology and Growth: Metabolomics helps study the metabolic pathways and compounds involved in plant growth, development, and responses to environmental cues. It reveals how plants adapt to changing conditions and helps optimize growth conditions.
Stress Responses: Metabolomics uncovers changes in metabolite profiles under abiotic stressors (drought, heat, cold) and biotic stressors (pathogens, pests). Understanding these responses can lead to the development of stress-tolerant crops.
Crop Improvement: Metabolomics aids in identifying traits associated with desirable crop characteristics, such as nutritional content, flavor, and aroma. It assists in breeding programs for developing improved crop varieties.
Secondary Metabolites: Study of secondary metabolites (e.g., alkaloids, flavonoids) provides insights into plant defense mechanisms, antimicrobial compounds, and potential pharmaceutical applications.
Pharmacology and Natural Products: Plant metabolomics helps identify and characterize bioactive compounds with potential pharmaceutical applications, contributing to drug discovery.
Food Quality and Safety: Metabolomics assesses food quality, authenticity, and safety by detecting metabolites related to freshness, nutritional value, and contaminants.
Nutritional Studies: Metabolomics analyzes nutritional content, bioactive compounds, and potential health benefits of plant-derived foods and supplements.
Environmental Monitoring: Metabolomics can indicate the health of ecosystems by studying the metabolite profiles of plants in response to pollution, climate change, and other environmental factors.
Biotechnology and Synthetic Biology: Metabolomics guides the metabolic engineering of plants for enhanced production of biofuels, industrial chemicals, and pharmaceuticals.
Plant-Microbe Interactions: Metabolomics reveals metabolic changes in plants during interactions with beneficial or pathogenic microbes, aiding in understanding symbiotic relationships and disease mechanisms.
Phytoremediation: Metabolomics helps identify plants that can remediate pollutants from soil and water by analyzing their metabolite responses.
Plant-Microbiome Interactions: Metabolomics studies the impact of the plant microbiome on host plant metabolism and health.
Sample Requirements for Plants Metabolomics Assay
| Sample Types |
Minimum Sample Size |
| Plant Samples |
Roots, stems and leaves, floral parts, fruits/seeds, rhizomes, buds/tender leaves, tissue sections, pollen, bark, trunk/wood, resin/gum, resin acids, seedlings/young plants, rhizosphere soil, root exudates. |
50 mg - 1 g |
Case 1. Metabolomics Reveals Altered Metabolite Profiles and Defense Strategies in Pathogen-Infected Rice Plants
Background:
Plants are in constant interaction with a diverse array of microorganisms throughout their lifecycle, encompassing both nonpathogenic and pathogenic entities. Investigating the relationships between plants and pathogens is of paramount importance due to the intricate dynamics involved and the wealth of metabolites implicated in these interactions. Metabolomics, a potent analytical technique, enables a holistic examination of the metabolites within biological systems, thereby offering a unique avenue to delve into the mechanisms underlying plant defense responses.
Samples:
The experimental framework involved the utilization of rice plants (Oryza sativa) as the model system. To emulate a pathogenic scenario, these rice plants were subjected to infection by the rice blast fungus (Magnaporthe oryzae). In parallel, control samples encompassed non-infected rice plants. The central objective was to unravel the alterations in metabolite composition arising from the imposition of pathogenic stress.
Methods:
Inoculation and Growth Conditions: Rice plants at specific developmental stages, specifically two or three leaf stages, were chosen for inoculation. Infection was facilitated by administering a spore suspension of Magnaporthe oryzae onto the rice foliage. Subsequent to inoculation, the plants were subjected to controlled growth conditions involving a humidified chamber and, subsequently, a growth chamber.
Sample Preparation: Following the prescribed growth period, both infected and control rice leaves were meticulously harvested. These plant tissues were then transformed into finely powdered form using liquid nitrogen and a mortar-pestle setup. To facilitate metabolite extraction, methanol was introduced into the powdered tissue, followed by vigorous vortexing and sonicating. The resulting supernatant was subjected to filtration using a 0.2 μm pore syringe filter. The filtrate, harboring the metabolites of interest, underwent a controlled evaporation process and was stored at sub-zero temperatures.
UPLC-QTOF-MS Analysis: The analytical facet involved an advanced UPLC-QTOF-MS system, combining ultra-high performance liquid chromatography (UPLC) with quadrupole time-of-flight mass spectrometry (QTOF-MS). This instrumental setup allowed for precise separation and identification of metabolites. The process commenced with the acquisition of mass features utilizing the MZmine software. These features were subsequently subjected to meticulous analysis and interpretation.
Feature Finding: Mass feature detection entailed a multi-step process. It encompassed cropping the mzML files, followed by mass detection and chromatogram deconvolution. The deconvoluted peaks were further processed to align the peaks, culminating in the generation of a comprehensive peak table.
Statistical Data Analysis: The acquired data underwent an array of statistical analyses. This included fold change analysis to ascertain differential metabolite levels, t-tests for significance assessment, and multivariate analyses such as PCA and PLS-DA to decipher patterns within the data. Heatmap clustering was additionally employed to elucidate relationships among the metabolite profiles.
Annotation and Pathway Analysis: The identification of metabolites involved comparing the observed mass and fragmentation patterns with established spectral databases. Pathway analysis, facilitated by the KEGG pathway database, enabled the exploration of the underlying biological processes connected with the identified metabolites.
Results
The investigation unearthed substantial shifts in the metabolite profiles of pathogen-infected rice plants in comparison to the controls. A total of twelve metabolites were earmarked as notably induced in the infected rice plants, with nine of them being successfully annotated and characterized. These metabolites were associated with diverse plant defense mechanisms, spanning amino acid metabolism, cytokinin biosynthesis, and lipid metabolism. A particularly intriguing discovery was the heightened presence of prostanoids and octadecanoids, derived from lipid metabolism, indicating their pivotal role in the plant's response to pathogenic invasion. This comprehensive analysis not only deepened our comprehension of plant-pathogen interactions but also underscored the potency of metabolomics in unraveling the intricacies of plant defense strategies.
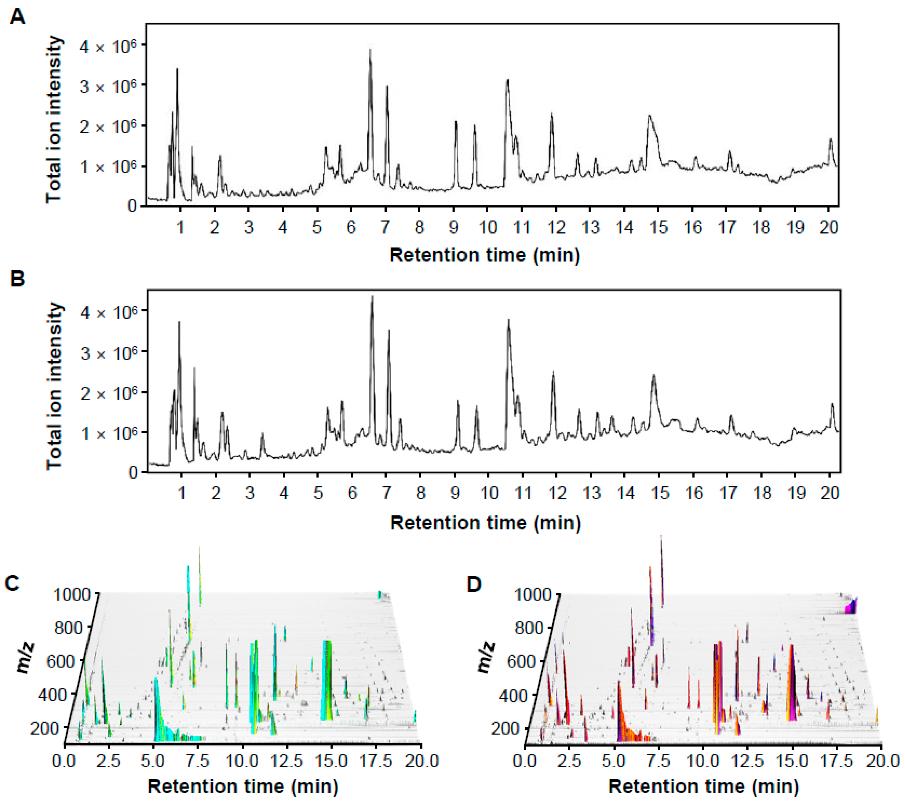 Representative LC-MS chromatographic profiling of control and pathogen-infected rice in positive ion mode.
Representative LC-MS chromatographic profiling of control and pathogen-infected rice in positive ion mode.
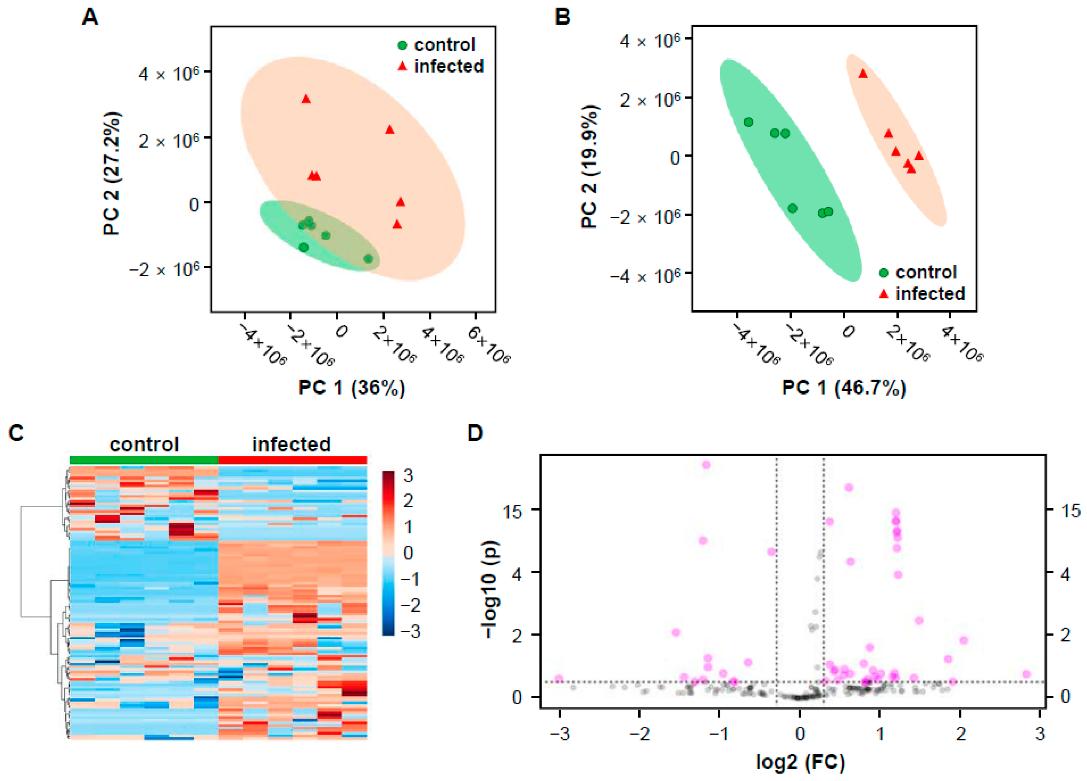 Multivariate statistical analysis of control and infected rice groups based on the mass features that generated by MZmine 2.53.
Multivariate statistical analysis of control and infected rice groups based on the mass features that generated by MZmine 2.53.
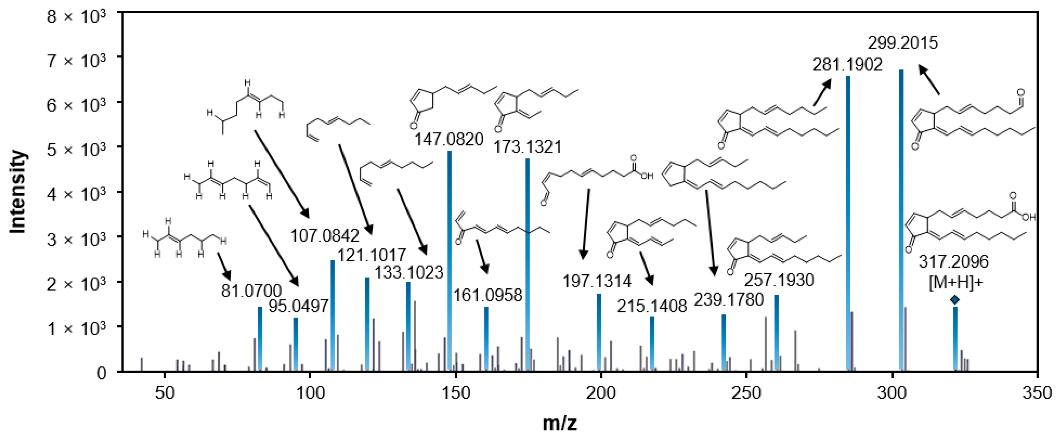 LC-MS/MS fragmentation pattern of 15-deoxy-Δ12,14-prostaglandin J2.
LC-MS/MS fragmentation pattern of 15-deoxy-Δ12,14-prostaglandin J2.
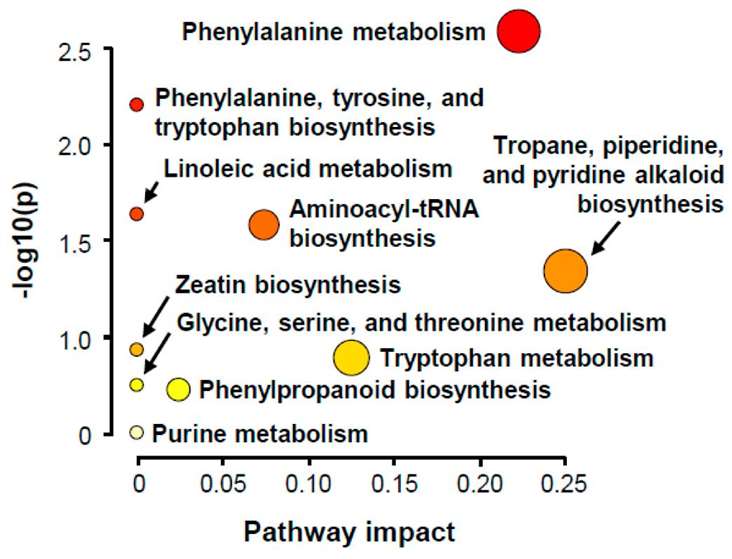 Summary plot of KEGG pathway analysis
Summary plot of KEGG pathway analysis
Reference
- Oh, Mira, et al. "Application of UPLC-QTOF-MS based untargeted metabolomics in identification of metabolites induced in pathogen-infected rice." Plants 10.2 (2021): 213.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service Representative LC-MS chromatographic profiling of control and pathogen-infected rice in positive ion mode.
Representative LC-MS chromatographic profiling of control and pathogen-infected rice in positive ion mode. Multivariate statistical analysis of control and infected rice groups based on the mass features that generated by MZmine 2.53.
Multivariate statistical analysis of control and infected rice groups based on the mass features that generated by MZmine 2.53. LC-MS/MS fragmentation pattern of 15-deoxy-Δ12,14-prostaglandin J2.
LC-MS/MS fragmentation pattern of 15-deoxy-Δ12,14-prostaglandin J2. Summary plot of KEGG pathway analysis
Summary plot of KEGG pathway analysis

