What is Aspartate Metabolism?
Aspartate, also recognized as aspartic acid, acts as a link between amino acid and sugar metabolism pathways, facilitating the interchange of metabolic intermediates between these interconnected networks. Its significance extends beyond flavor enhancement to encompass pivotal functions in nitrogen metabolism, where it serves as a precursor for the synthesis of other amino acids, purines, and pyrimidines. Moreover, aspartate serves as a vital nitrogen storage form and participates in nitrogen assimilation and remobilization processes within plants.
Aspartate metabolism encompasses the biochemical processes involving the utilization, synthesis, and interconversion of aspartate. By elucidating the key pathways and regulatory mechanisms of aspartate metabolism, researchers can uncover novel targets for crop improvement, biofortification strategies, and sustainable agricultural practices. Our service facilitates scientific breeding efforts directed towards improving the nutritional content of fruits or seeds, and implementing strategies for tolerance against both biotic and abiotic stresses, including insect resistance, salt tolerance, and heat tolerance.
Aspartate Metabolism Analysis Service by Creative Proteomics
Aspartate Metabolism Metabolite Profiling: Within Creative Proteomics, our wide array of services includes in-depth metabolite profiling, providing thorough quantitative assessments of aspartate and its derivatives across various plant samples. Leveraging state-of-the-art analytical platforms, we enable efficient screening of metabolites on a large scale, facilitating accurate detection of biomarkers and metabolic pathways linked to aspartate metabolism.
Aspartate Metabolism Correlation Analysis: Expanding our service portfolio, we delve into gene expression analysis, with a focus on the coordinated expression of genes involved in pathways associated with aspartate metabolism. This encompasses various processes, including amino acid biosynthesis and the synthesis of tricarboxylic acid (TCA) cycle intermediates. Through the utilization of advanced transcriptomic profiling techniques, we pinpoint genes participating in crucial enzymatic reactions within the aspartate metabolic network.
Aspartate Metabolism Metabolic Flux Analysis: Employing sophisticated isotopic labeling methods, we track the movement of carbon and nitrogen within aspartate metabolic pathways, enabling precise quantification of metabolic fluxes and contributions from different pathways.
Enzyme Activity Assays: We also assay the activity of enzymes crucial to aspartate metabolism, such as aspartate transaminase and glutamate synthase. These assessments provide insights into metabolic flux and enzyme kinetics, aiding in the comprehensive understanding of aspartate metabolic processes.
Techniques and Instrumentation for Aspartate Metabolism Analysis
Liquid Chromatography (LC): At Creative Proteomics's Aspartate Metabolism Analysis Service, liquid chromatography (LC) stands as a fundamental technique, pivotal for the isolation and purification of aspartate and its metabolites ahead of mass spectrometry examination. Utilizing our cutting-edge LC systems (Vanquish H), we guarantee exceptional chromatographic resolution and efficacy, streamlining the accurate quantification and recognition of desired analytes within intricate biological samples. Tailoring chromatographic parameters to specific requirements, we enhance the isolation of target analytes from potential interfering substances, thus securing precision and dependability in our findings.
Mass Spectrometry (MS): At Creative Proteomics, mass spectrometry (MS) holds a crucial position in the identification and quantification of aspartate and its derivatives, boasting unmatched sensitivity and specificity. Our approach involves the utilization of state-of-the-art MS platforms like the QE and Thermo Q-Exactive instruments, renowned for their high resolution and accuracy. Leveraging these cutting-edge tools, we ensure precise detection and quantification of compounds associated with aspartate across a wide range of plant specimens.
LC-MS/MS Method Establishment and Optimization: Creative Proteomics employs LC-MS/MS methods for targeted quantification of aspartate and its metabolites.Our approach involves two critical aspects:
(1) Standard Curve
We construct standard curves utilizing reference standards of aspartate and its related compounds. This enables us to quantitatively determine the target compounds in biological samples. Through meticulous calibration procedures, we ensure the linearity, accuracy, and reproducibility of the standard curves, facilitating precise quantification of aspartate metabolites across a broad concentration range.
(2) Multiple Reaction Monitoring (MRM) Quantification
Creative Proteomics implements MRM quantification for the targeted analysis of aspartate and aspartate metabolites. By selecting suitable precursor and product ion pairs for each metabolite, we optimize MRM transitions to enhance detection efficiency and minimize interference from background noise.
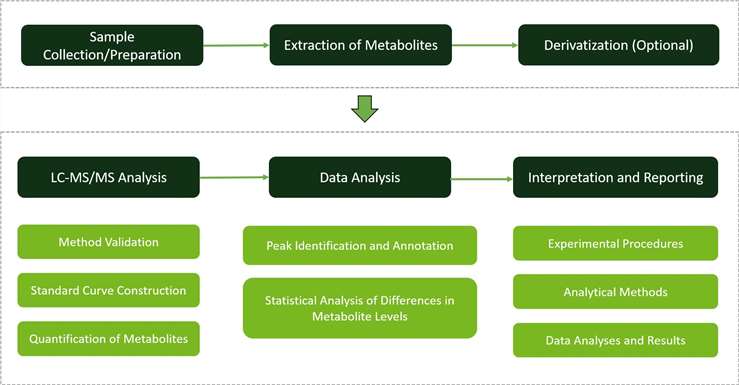 Workflow of Aspartate Metabolism Analysis Service
Workflow of Aspartate Metabolism Analysis Service
Why Choose Us?
- Targeted Metabolomics Approach: Creative Proteomics employs metabolomics approach specifically tailored to analyze the metabolism of aspartate and its derivatives. This focused methodology ensures high precision and sensitivity in detecting and quantifying aspartate-related metabolites in complex biological samples.
- Comprehensive Coverage: Creative Proteomics's sample preparation protocol cover a wide range of plant tissues and sample types, including roots, stems, leaves, fruits, and seeds, ensuring comprehensive analysis of aspartate metabolism across various developmental stages and environmental conditions.
- Personalized Sample Processing: We utilize state-of-the-art analytical techniques LC-MS/MS to achieve comprehensive profiling of aspartate metabolism. Advanced methods enable accurate identification and quantification of metabolites, even at low concentrations, facilitating in-depth analysis of metabolic pathways.
- Robust Quality Control Measures: Our service implements stringent quality control measures at every stage of the analysis process. This includes the use of internal standards, calibration curves, and quality control samples to ensure the accuracy, reliability, and reproducibility of our data.
- Expert Data Analysis and Interpretation: In addition to providing metabolism analytical services, we offer expert data analysis and interpretation. We use a parallel method of human and machine comparison to analyze the preprocessed data and provide customized analysis report services based on research needs.
Applications of Aspartate Metabolism Analysis
Improving Crop Flavor: Exploration of aspartate metabolism contributes to understand and manipulate pathways crucial for flavor enhancement, elevating the taste and sensory appeal of specific crops.
Understanding Plant Nitrogen Utilization: Isotopic labeling enables the investigation of nitrogen metabolism intricacies, revealing how plants assimilate, distribute, and utilize nitrogen resources.
Unveiling Stress Responses: Analyzing aspartate metabolism dynamics deciphers plant stress response molecular mechanisms, aiding in developing stress-tolerant crop varieties via genetic manipulation or breeding strategies.
Sample Requirements for Aspartate Metabolism Assay
| Plant Sample Type |
Minimum Quantity Required(fresh weight) |
Storage Conditions |
Additional Notes |
| Roots,Stems,Leaves,Fruits |
>200 mg |
Quick freeze in liquid nitrogen, Store at -80°C |
- |
| Seeds |
>200 mg |
Include seeds if applicable |
Each experimental treatment should have more than 6 biological replicates to ensure robust statistical analysis and reliable interpretation of results.
For other sample types not listed above, such as flowers or whole plants, please consult our technical support or sales team for specific requirements and recommendations.
Case. Identification of key genes contributing to amino acid biosynthesis in Torreya grandis using transcriptome and metabolome analysis
Background:
Torreya grandis, renowned for its economic and nutritional value, particularly in its kernels, serves as the focal point of investigation. The kernels of different development stages vary enormously in their amino acids content.
The molecular mechanisms underlying amino acid biosynthesis in T. grandis remain inadequately understood. Transcriptome and metabolome analyses present a holistic approach to unraveling these pathways, facilitating the identification of candidate genes and regulatory factors.
Samples:
Plant materials from three T. grandis cv. 'Merrilli' grafting trees at different seed development stages were collected.
Plants underwent a 28-day acclimation period in the greenhouse, with controlled temperature and lighting times, and regular maintenance.
After collection, kernels were frozen in liquid nitrogen and stored at -80°C. Each biological replicate comprised 10 seeds, with three replicates per experiment for RNA sequencing, metabolite profiling, and physiological assays.
Technical methods procedure:
Metabolite extraction involved grinding samples in liquid nitrogen, followed by dissolution in methanol.
LC-ESI-MS/MS analysis was conducted to analyze metabolites and widely targeted metabolite profiling reveals the metabolites in Torreya grandis seed maturation.
Differential gene expression was analyzed using DESeq2 software, and KEGG pathway enrichment analysis was conducted.
Subcellular localization was determined using GFP fusion proteins, and transgenic Arabidopsis lines were generated to assess amino acid content.
Dual luciferase assays were used to investigate transcription factor activity on TgASA1 promoter.
Results
Amino acid content varied significantly during development, with 28 amino acids identified, including aspartate, which is a major component of the tricarboxylic acid cycle (TCA) pathway and amino acid biosynthesis.
Correlation analysis identified key genes involved in amino acid biosynthesis. Subcellular localization of key enzymes TgDAHP2 and TgASA1 confirmed chloroplast localization. Overexpression of TgDAHP2 and TgASA1 in Arabidopsis significantly increased amino acid content.
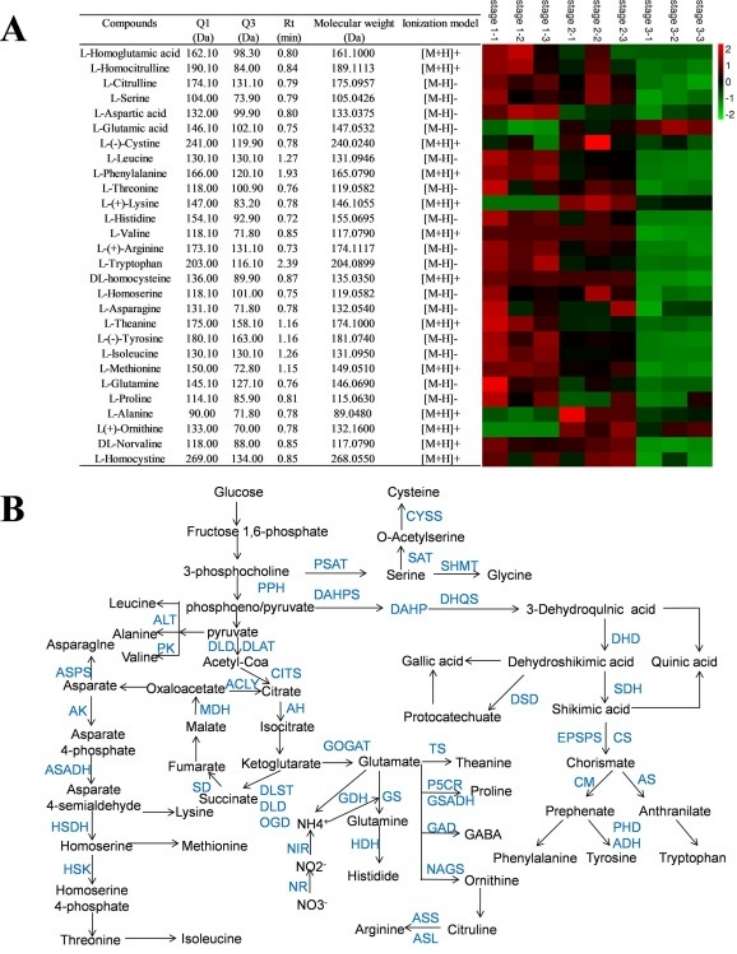 Fig 1. Gene expression and amino acids analysis for amino acid synthesis pathway of T. grandis. (A) The relative contents of amino acids in T. grandis kernels in the three mature stages. (B) Schematic diagram of the tricarboxylic acid cycle (TCA) pathway and amino acid biosynthesis in T. grandis.
Fig 1. Gene expression and amino acids analysis for amino acid synthesis pathway of T. grandis. (A) The relative contents of amino acids in T. grandis kernels in the three mature stages. (B) Schematic diagram of the tricarboxylic acid cycle (TCA) pathway and amino acid biosynthesis in T. grandis.
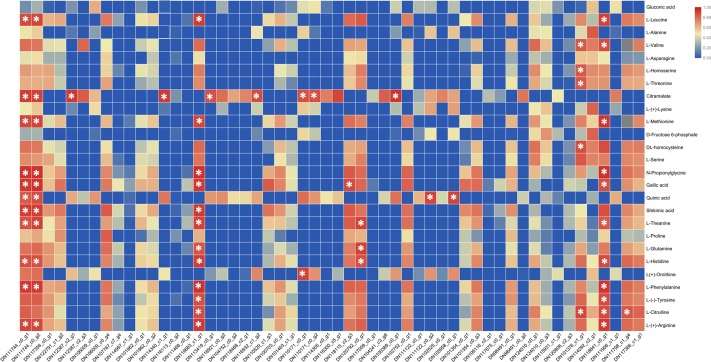 Fig 2. Pearson correlation analysis between amino acid coding genes and metabolites in amino acid synthesis pathway. The Pearson correlation analysis between the expression levels of the possible coding genes selected by the phylogenetic tree analysis and the metabolites on the amino acid synthesis pathway in the three mature stage of T. grandis seeds. The significant ones are marked with * (p < 0.05).
Fig 2. Pearson correlation analysis between amino acid coding genes and metabolites in amino acid synthesis pathway. The Pearson correlation analysis between the expression levels of the possible coding genes selected by the phylogenetic tree analysis and the metabolites on the amino acid synthesis pathway in the three mature stage of T. grandis seeds. The significant ones are marked with * (p < 0.05).
Reference
- Lou, H. (2022). "Identification of Key Genes Contributing to Amino Acid Biosynthesis in Torreya Grandis Using Transcriptome and Metabolome Analysis." Food Chemistry 379 (2022): 132078.


 Workflow of Aspartate Metabolism Analysis Service
Workflow of Aspartate Metabolism Analysis Service Fig 1. Gene expression and amino acids analysis for amino acid synthesis pathway of T. grandis. (A) The relative contents of amino acids in T. grandis kernels in the three mature stages. (B) Schematic diagram of the tricarboxylic acid cycle (TCA) pathway and amino acid biosynthesis in T. grandis.
Fig 1. Gene expression and amino acids analysis for amino acid synthesis pathway of T. grandis. (A) The relative contents of amino acids in T. grandis kernels in the three mature stages. (B) Schematic diagram of the tricarboxylic acid cycle (TCA) pathway and amino acid biosynthesis in T. grandis. Fig 2. Pearson correlation analysis between amino acid coding genes and metabolites in amino acid synthesis pathway. The Pearson correlation analysis between the expression levels of the possible coding genes selected by the phylogenetic tree analysis and the metabolites on the amino acid synthesis pathway in the three mature stage of T. grandis seeds. The significant ones are marked with * (p < 0.05).
Fig 2. Pearson correlation analysis between amino acid coding genes and metabolites in amino acid synthesis pathway. The Pearson correlation analysis between the expression levels of the possible coding genes selected by the phylogenetic tree analysis and the metabolites on the amino acid synthesis pathway in the three mature stage of T. grandis seeds. The significant ones are marked with * (p < 0.05).

