What is Lysine Metabolism?
Lysine, an essential amino acid, plays a multifaceted role in plant metabolism, influencing growth, development, and stress responses. Synthesized via the aspartate pathway, lysine is integral to protein synthesis and can be catabolized into Acetyl-CoA, a critical component of the tricarboxylic acid (TCA) cycle, thereby contributing to cellular energy production. The regulation of lysine metabolism affects the levels of other amino acids such as threonine, methionine, and isoleucine, underscoring a tightly controlled metabolic network essential for maintaining metabolic homeostasis.
Lysine metabolism involves the synthesis, utilization, and breakdown of lysine, and Lysine-derived metabolites like pipecolic acid play significant roles in plant stress responses and immunity. Creative Proteomics's Lysine Metabolism Analysis Service offers detailed insights into lysine pathways, aiding the development of nutritionally enhanced, disease-resistant, and stress-tolerant crops through advanced profiling and gene expression analysis.
Lysine Metabolism Analysis Service by Creative Proteomics
Lysine Metabolism Metabolite Profiling: At Creative Proteomics, we leverage advanced mass spectrometry and chromatography methods to quantify lysine and its metabolic intermediates precisely. Our techniques enable the accurate detection and quantification of key compounds like pipecolic acid and N-hydroxypipecolic acid, facilitating comprehensive metabolic profiling.
Lysine Metabolism Pathway Mapping: Sophisticated bioinformatics tools are employed for elucidating the intricate network of lysine metabolism. By visualizing metabolic pathways, we identify potential regulatory nodes and interaction points with other metabolic processes, offering a holistic understanding of lysine metabolism in plants.
Enzyme Activity Assays Related to Lysine Metabolic Pathway: Determining the catalytic activity of key enzymes in the lysine metabolic pathway is critically important. Our thorough analysis provides valuable insights into the dynamic regulation of lysine metabolism, highlighting key enzymatic functions and interactions within plant systems.
Customized Solutions for Lysine Metabolism: Acknowledging the unique requirements of different plant species and research objectives, we provide bespoke analysis packages tailored to your needs. Whether for fundamental research elucidating lysine metabolic pathways or applied agricultural biotechnology enhancing lysine content in crops, our customized solutions cater to diverse research goals and applications.
Techniques and Instrumentation for Lysine Metabolism Analysis
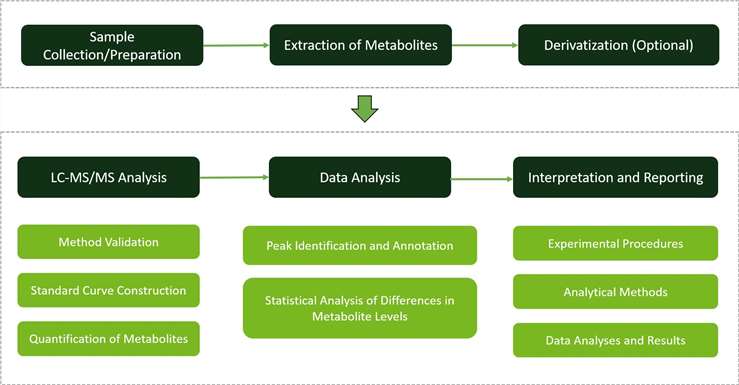
Liquid Chromatography (LC): We employ advanced liquid chromatography systems to separate and purify lysine and its metabolites from complex biological samples. Our LC systems (Vanquish H) offer exceptional resolution and efficiency, ensuring accurate quantification of lysine and its derivatives.
Mass Spectrometry (MS): Mass spectrometry is integral to our analysis, enabling precise identification and quantification of lysine and its metabolites. We utilize state-of-the-art mass spectrometry platforms (Q-Exactive) renowned for their sensitivity and specificity, ensuring reliable results across a wide range of plant samples.
LC-MS/MS Method Establishment and Optimization: Our approach involves the establishment and optimization of LC-MS/MS methods for targeted quantification of lysine and its metabolites. We construct standard curves using reference standards of lysine and related compounds to ensure accurate quantification over a broad concentration range. Multiple Reaction Monitoring (MRM) quantification is employed to enhance detection efficiency and minimize background noise, ensuring precise analysis of lysine metabolism.
Why Choose Us?
- High Sensitivity and Specificity: Our cutting-edge instruments offer unparalleled sensitivity and specificity, enabling the detection of lysine and its derivatives at extremely low concentrations. This ensures the acquisition of precise and dependable data, even from complex biological samples.
- Comprehensive Profiling: Creative Proteomics's Lysine Metabolism Analysis Service offers an extensive profiling of lysine and its metabolic intermediates. This allows for precise quantification and identification of key metabolites, ensuring a thorough understanding of lysine metabolism in plant systems.
- Enhanced Resolution and Efficiency: Our UPLC system delivers exceptional chromatographic resolution and efficiency. This enhances the separation and purification of lysine and its metabolites, ensuring that our analysis is both accurate and dependable. Tailored chromatographic parameters further optimize the isolation of target analytes from interfering substances.
- High-Throughput Capability: Creative Proteomics's advanced analytical platforms enable high-throughput screening of lysine and its metabolites. This capability ensures that we can handle extensive sample sets efficiently, delivering results promptly without compromising on quality.
- Sophisticated Data Analysis: We possess numerous reference standards, and by combining these with the MRM method of LC-MS/MS, we ensure the integrity, accuracy, and reproducibility of our measurements. Advanced bioinformatics tools help in constructing detailed metabolic pathways, visualizing interactions, and identifying regulatory nodes.
Applications of Lysine Metabolism Analysis
Nutritional Enhancement in Crops: Leveraging Creative Proteomics's Lysine Metabolism Analysis Service allows researchers to enhance lysine levels in crops, thus addressing deficiencies in essential amino acids. This results in biofortified foods, which can significantly improve global food security and public health.
Improving Disease Resistance: Our service offers comprehensive insights into the impact of lysine metabolism on plant immune responses. This knowledge is instrumental in engineering crops with increased resistance to pathogens, thereby reducing dependence on chemical pesticides and fostering sustainable farming practices.
Enhancing Stress Tolerance: Creative Proteomics's service aids researchers in comprehending the role of lysine in plant stress responses, such as drought and salinity. By examining metabolite profiles and gene expression, scientists can develop crops that sustain high yields even under challenging environmental conditions.
Advancing Genetic Engineering: Our detailed profiling of lysine metabolic pathways supports progress in genetic engineering. Researchers can pinpoint key regulatory genes and enzymes, facilitating the creation of genetically modified crops with specific traits like faster growth rates or increased biomass production.
Sample Requirements for Lysine Metabolism Assay
| Plant Sample Type |
Minimum Quantity Required (fresh weight) |
Storage Conditions |
Additional Notes |
| Roots,Stems,Leaves,Fruits |
>200 mg |
Quick freeze in liquid nitrogen, Store at -80°C |
- |
| Seeds |
>200 mg |
Include seeds if applicable |
Each experimental treatment should have more than 6 biological replicates to ensure robust statistical analysis and reliable interpretation of results.
For other sample types not listed above, such as flowers or whole plants, please consult our technical support or sales team for specific requirements and recommendations.
Case . Identification of key taste components in loquat using widely targeted metabolomics
Background:
This study investigates the taste differences between white-fleshed and yellow-fleshed loquats using widely targeted metabolomics. White-fleshed loquats are known for their superior taste compared to yellow-fleshed varieties.
Utilizing LC-MS/MS, the research aims to identify key metabolites and pathways responsible for these taste differences, providing insights into the molecular basis of loquat flavor profiles.
Samples:
In this study, two loquat cultivars, 'Baiyu' (white-fleshed) and 'Zaozhong No. 6' (yellow-fleshed), were utilized.
Mature fruits were harvested, peeled, and pooled to create biological samples for each cultivar, with a minimum of three biological samples per cultivar being analyzed.
The sample preparation involved freeze-drying and finely grinding the loquat flesh, followed by extraction with 70% methanol. The mixture was centrifuged, and the supernatant was collected for analysis.
Technical methods procedure:
Quality control samples were prepared and analyzed alongside the fruit samples to ensure reproducibility.
loquat fruit extracts were analyzed using an LC-ESI-MS/MS system, employing a specific HPLC column and gradient.
Mass spectrometry was conducted with a triple quadrupole-linear ion trap system, optimizing parameters for ion source operation, collision energy, and declustering potential for each multiple reaction monitoring (MRM) transition, ensuring precise metabolite identification and quantification.
Utilized MS2 spectral tag library for metabolite identification, facilitating downstream statistical and pathway analyses. Utilized PCA and OPLS-DA for dimensionality reduction and pattern recognition, ensuring model robustness through permutation testing. Mapped metabolites to KEGG database, performed enrichment analysis via MSEA, unveiling metabolic alterations under experimental conditions.
Results
Differential metabolites were categorized into various classes, primarily carbohydrates, organic acids, amino acids, phenolics, and lipids. KEGG pathway analysis revealed differences in specialized plant pathways, notably flavonoid biosynthesis.
Carbohydrates and organic acids significantly differed between cultivars, with 'Baiyu' exhibiting higher concentrations, potentially explaining its sweeter taste. Amino acid composition variations further contributed to taste differences. Three of these (L-glutamine, L-citrulline and L-(+)-lysine) were significantly and differentially accumulated between the cultivars.
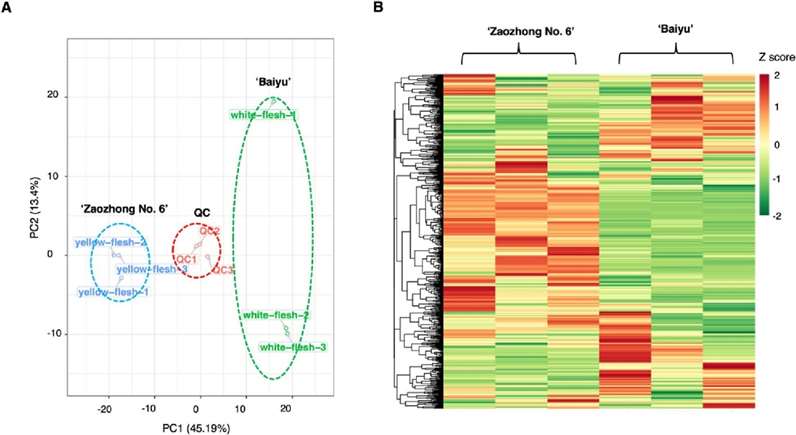 Fig 1. Differential fruit chemotype between 'Baiyu' and 'Zaozhong No. 6'. (A) PCA analysis of metabolites identified from 'Baiyu' and 'Zaozhong No. 6'. Equal volumes of 'Baiyu' and 'Zaozhong No. 6' fruit samples were mixed for use as a quality control (QC). (B) Cluster analysis of metabolites from samples of 'Baiyu' and 'Zaozhong No. 6'. The colour indicates the level of accumulation of each metabolite, from low (green) to high (red). The Z-score represents the deviation from the mean by standard deviation units.
Fig 1. Differential fruit chemotype between 'Baiyu' and 'Zaozhong No. 6'. (A) PCA analysis of metabolites identified from 'Baiyu' and 'Zaozhong No. 6'. Equal volumes of 'Baiyu' and 'Zaozhong No. 6' fruit samples were mixed for use as a quality control (QC). (B) Cluster analysis of metabolites from samples of 'Baiyu' and 'Zaozhong No. 6'. The colour indicates the level of accumulation of each metabolite, from low (green) to high (red). The Z-score represents the deviation from the mean by standard deviation units.
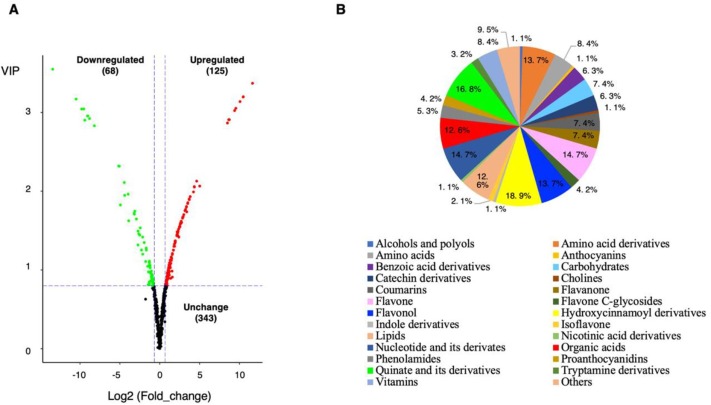 Fig 2. Differentially accumulating metabolites between 'Baiyu' and 'Zaozhong No. 6'. (A)Volcano plot of the 536 metabolites identified. Differential metaboliteswere defined as metabolites with fold change ≥ 1.6 or ≤ 0.625 in 'Zaozhong No. 6' compared to 'Baiyu'. A threshold of VIP ≥ 0.8 was used to separate differential metabolites from unchanged metabolites. (B) Pie chart depicting the biochemical categories of the differential metabolites identified between 'Baiyu' and 'Zaozhong No. 6'.
Fig 2. Differentially accumulating metabolites between 'Baiyu' and 'Zaozhong No. 6'. (A)Volcano plot of the 536 metabolites identified. Differential metaboliteswere defined as metabolites with fold change ≥ 1.6 or ≤ 0.625 in 'Zaozhong No. 6' compared to 'Baiyu'. A threshold of VIP ≥ 0.8 was used to separate differential metabolites from unchanged metabolites. (B) Pie chart depicting the biochemical categories of the differential metabolites identified between 'Baiyu' and 'Zaozhong No. 6'.
Reference
- Zou, S. (2020). " Identification of key taste components in loquat using widely targeted metabolomics." Food Chemistry 323, 126822.



 Fig 1. Differential fruit chemotype between 'Baiyu' and 'Zaozhong No. 6'. (A) PCA analysis of metabolites identified from 'Baiyu' and 'Zaozhong No. 6'. Equal volumes of 'Baiyu' and 'Zaozhong No. 6' fruit samples were mixed for use as a quality control (QC). (B) Cluster analysis of metabolites from samples of 'Baiyu' and 'Zaozhong No. 6'. The colour indicates the level of accumulation of each metabolite, from low (green) to high (red). The Z-score represents the deviation from the mean by standard deviation units.
Fig 1. Differential fruit chemotype between 'Baiyu' and 'Zaozhong No. 6'. (A) PCA analysis of metabolites identified from 'Baiyu' and 'Zaozhong No. 6'. Equal volumes of 'Baiyu' and 'Zaozhong No. 6' fruit samples were mixed for use as a quality control (QC). (B) Cluster analysis of metabolites from samples of 'Baiyu' and 'Zaozhong No. 6'. The colour indicates the level of accumulation of each metabolite, from low (green) to high (red). The Z-score represents the deviation from the mean by standard deviation units. Fig 2. Differentially accumulating metabolites between 'Baiyu' and 'Zaozhong No. 6'. (A)Volcano plot of the 536 metabolites identified. Differential metaboliteswere defined as metabolites with fold change ≥ 1.6 or ≤ 0.625 in 'Zaozhong No. 6' compared to 'Baiyu'. A threshold of VIP ≥ 0.8 was used to separate differential metabolites from unchanged metabolites. (B) Pie chart depicting the biochemical categories of the differential metabolites identified between 'Baiyu' and 'Zaozhong No. 6'.
Fig 2. Differentially accumulating metabolites between 'Baiyu' and 'Zaozhong No. 6'. (A)Volcano plot of the 536 metabolites identified. Differential metaboliteswere defined as metabolites with fold change ≥ 1.6 or ≤ 0.625 in 'Zaozhong No. 6' compared to 'Baiyu'. A threshold of VIP ≥ 0.8 was used to separate differential metabolites from unchanged metabolites. (B) Pie chart depicting the biochemical categories of the differential metabolites identified between 'Baiyu' and 'Zaozhong No. 6'.

