Oryza sativa Metabolomics
Oryza sativa metabolomics involves the comprehensive study of metabolites within the biological system of Oryza sativa, commonly known as rice. Metabolomics, a subfield of omics sciences, focuses on analyzing the small molecules—metabolites—that are integral to various cellular processes. These metabolites provide insights into the biochemical status and physiological responses of the organism. In the case of Oryza sativa, a pivotal cereal crop globally, metabolomics plays a crucial role in understanding its growth, development, and reactions to diverse environmental conditions.

What Oryza sativa Metabolomics Can Analyze?
Plant Growth and Development: Metabolites play essential roles in regulating growth and development, including processes like cell division, differentiation, and nutrient allocation. Metabolomics enables the identification of key metabolites associated with these processes, facilitating the development of strategies to increase yield and improve nutritional content.
Stress Responses: Oryza sativa faces a range of challenges, from pathogen attacks to drought, salinity, and temperature fluctuations. Metabolomics reveals the metabolic changes that occur under stress conditions, enabling the identification of stress-responsive metabolites and pathways. This knowledge guides the breeding of stress-tolerant Oryza sativa varieties.
Nutritional Quality: Metabolomics uncovers the metabolites responsible for nutritional attributes in Oryza sativa, such as vitamins, minerals, and secondary metabolites. This information is critical for developing nutrient-rich rice varieties to address nutritional deficiencies.
Crop Improvement: By identifying metabolic pathways that contribute to desirable traits, such as disease resistance or enhanced photosynthesis, researchers can use targeted genetic modification or breeding techniques to develop improved Oryza sativa varieties.
Agronomic Practices: Metabolomics assists in optimizing agricultural practices by providing insights into how factors like fertilization and irrigation impact Oryza sativa metabolism and overall yield.
Oryza sativa Metabolomics Analysis at Creative Proteomics
Targeted Metabolite Profiling:
Analyzing specific classes of metabolites or individual compounds to quantify their levels in Oryza sativa samples. This involves accurate measurements using LC-MS or GC-MS platforms.
Untargeted Metabolomics:
Conducting comprehensive analysis of the complete metabolome in Oryza sativa samples using advanced LC-MS or GC-MS techniques. This includes identifying and quantifying a wide range of metabolites.
Metabolic Pathway Analysis:
Mapping metabolites onto relevant metabolic pathways to understand the interconnected biochemical processes in Oryza sativa. This reveals insights into how different pathways are influenced by various growth conditions or stressors.
Biomarker Discovery:
Identifying potential biomarkers associated with specific traits, developmental stages, or stress responses in Oryza sativa. This involves pinpointing metabolites indicative of physiological changes.
Isotope Labeling Studies (SIRM):
Studying metabolic fluxes and pathways by incorporating stable isotopes into Oryza sativa samples. This enables the tracing of isotopically labeled metabolites and quantifying metabolic fluxes using LC-MS or GC-MS.
Comparative Metabolomics:
Comparing the metabolite profiles of different Oryza sativa samples to identify variations under different conditions or treatments. This uncovers how genetic modifications, environmental factors, or cultural practices affect rice metabolism.
Multi-omics Integration:
Integrating metabolomics data with genomics, transcriptomics, and proteomics data to achieve a holistic understanding of Oryza sativa biology. This reveals correlations between gene expression, protein levels, and metabolite changes.
Data Interpretation and Visualization:
Using specialized software tools to process, analyze, and visualize complex metabolomics data. This offers insights into significant metabolic changes, pathway enrichment, and metabolite interactions.
Customized Metabolomics Solutions:
Collaborating closely with researchers to design tailored metabolomics experiments addressing specific inquiries related to Oryza sativa. This involves providing expertise and guidance to optimize experimental designs and data analysis strategies.
Oryza sativa Metabolomics Analysis Techniques
Liquid Chromatography-Mass Spectrometry (LC-MS): Thermo Scientific™ Q Exactive™ Plus is utilized for both targeted and untargeted metabolite analysis in Oryza sativa. This technique combines liquid chromatography separation with high-resolution mass spectrometry detection, enabling comprehensive identification and quantification of metabolites.
Gas Chromatography-Mass Spectrometry (GC-MS): The Agilent 7890A GC System with 5977B GC/MSD is employed for profiling volatile metabolites in Oryza sativa. This method is particularly suited for compounds with lower molecular weights, offering insights into the volatile metabolic components of rice samples.
High-Resolution Mass Spectrometry (HRMS): The Bruker maXis II ETD QTOF Mass Spectrometer is used for untargeted metabolomics analysis in Oryza sativa. HRMS provides high-resolution and accurate mass measurements, facilitating the identification of diverse metabolites, including those with complex structures.
Triple Quadrupole Mass Spectrometry (QQQ-MS): The Agilent 6495 Triple Quadrupole LC/MS System enables precise targeted metabolomics in Oryza sativa. It quantifies specific metabolites, contributing to insights into metabolic pathways and dynamics within rice samples.
Orbitrap Mass Spectrometry: Thermo Scientific™ Orbitrap Fusion™ Tribrid™ Mass Spectrometer offers high-resolution analysis for Oryza sativa metabolites. This versatile system integrates multiple analyzers, such as quadrupole, ion trap, and Orbitrap, providing comprehensive qualitative and quantitative capabilities.
Quadrupole Time-of-Flight Mass Spectrometry (QTOF-MS): The Agilent 6546 LC/Q-TOF MS System is dedicated to metabolite identification in Oryza sativa. With high-resolution and fragmentation capabilities, QTOF-MS assists in determining metabolite structures present in rice samples.
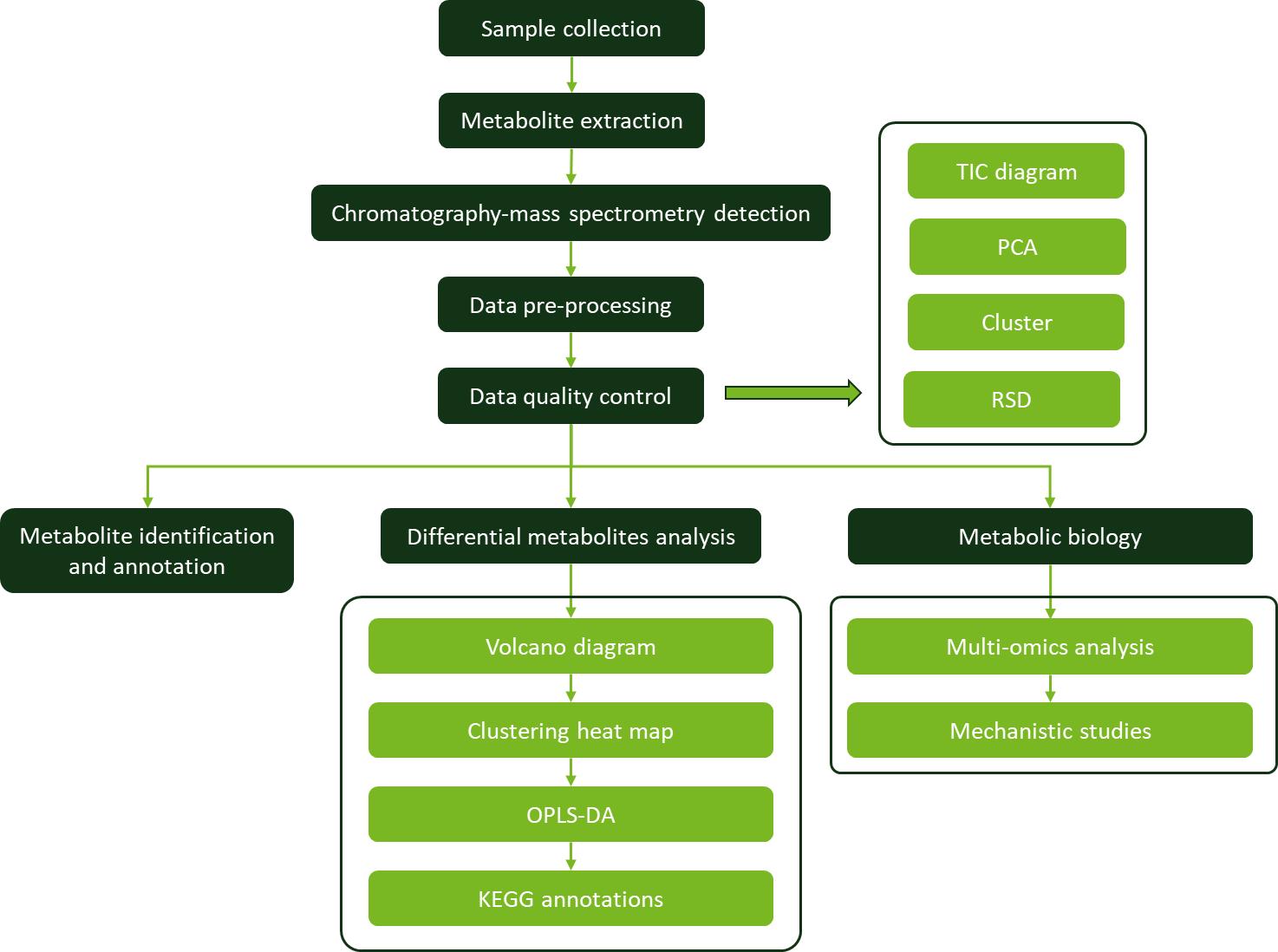 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
Sample Requirements for Oryza sativa Metabolomics
| Sample Type |
Growth Stage |
Biological Replicates |
Sample Quantity |
Extraction Solvent |
Storage Temperature |
| Seedlings |
Early growth stages |
≥ 3 |
100 mg (per replicate) |
Methanol or ethanol |
-80°C |
| Vegetative Stages |
Mid growth stages |
≥ 3 |
150 mg (per replicate) |
Acetonitrile |
-80°C |
| Reproductive Stages |
Flowering, seeding |
≥ 3 |
200 mg (per replicate) |
Mix of solvents |
-80°C |
| Time-Course Experiments |
Various |
≥ 3 time points |
Variable (per time point) |
Various |
-80°C |
Case 1. Metabolic Insights into Seed Storability Dynamics: Unveiling Sugar and Amino Acid Contributions in Hybrid Rice Cultivars
Background:
Seed storability is a critical aspect of crop conservation and agriculture. The viability and quality of seeds during storage are influenced by various chemical compositions and metabolic processes. Natural storage leads to irreversible seed deterioration, impacting germination capacity and seedling establishment.
Samples:
The study used two hybrid rice cultivars, BY and IIY, with distinct seed storability characteristics. These cultivars share the same male parent, reducing genetic background differences. Seeds were analyzed before and after a 24-month natural storage period.
Methods:
Metabolomic analysis was conducted using gas chromatography tandem mass spectrometry (GC-MS). Metabolites were extracted from both cultivars' dry seeds and analyzed comprehensively. The analysis included several steps:
Metabolite Extractions: Seeds were ground and extracted using methanol:chloroform:water solution, and the supernatant was collected.
GC-MS Analysis: Extracted metabolites were derivatized with MSTFA and analyzed using GC-MS. The chromatography conditions included temperature ramps and ion source settings.
Data Analysis: Chroma TOF 4.3X software was used for data analysis, including peak extraction, calibration, alignment, and baseline filtering. LECO-Fiehn Rtx5 database and NIST library were used for peak identification. Multivariate statistical analyses (PCA, OPLS-DA) and univariate statistical analysis (ANOVA) were performed to identify significant metabolite changes.
Results
Metabolic Profiling: GC-MS analyses revealed distinct compound peaks in the chromatograms of the two cultivars' seeds, especially in the retention time range of 17 to 26 minutes. A total of 90 metabolite peaks were detected, including sugars, amino acids, fatty acids, and others.
Multivariate Statistical Analysis: PCA and OPLS-DA analyses demonstrated separation between BY and IIY seeds. More metabolites significantly changed in IIY seeds during the 24-month storage, indicating higher metabolic dynamics in storage-sensitive seeds.
Changes in Sugar-Related Metabolites: Sugar-related compounds exhibited varied levels between cultivars before and after storage. Raffinose content was consistently higher in long-term storage-sensitive seeds. Other sugars like glucose, cellobiose, and glycerol-3-phosphate showed increased levels in short-term storage-sensitive seeds.
Changes in Amino Acid-Related Metabolites: Amino acids exhibited significant variation between cultivars, with higher levels in storage-sensitive seeds. Tyrosine, proline, valine, and other AAs showed notable increases in IIY seeds. Changes in AA levels correlated with protein degradation and oxidative stress responses.
In conclusion, the study explored the metabolic basis of seed storability using a comprehensive metabolomics approach. The analysis highlighted the roles of specific sugars and amino acids in seed storability and emphasized the metabolic dynamics in storage-sensitive seeds. The findings provide valuable insights into seed deterioration during storage, with implications for crop conservation and seed quality management.
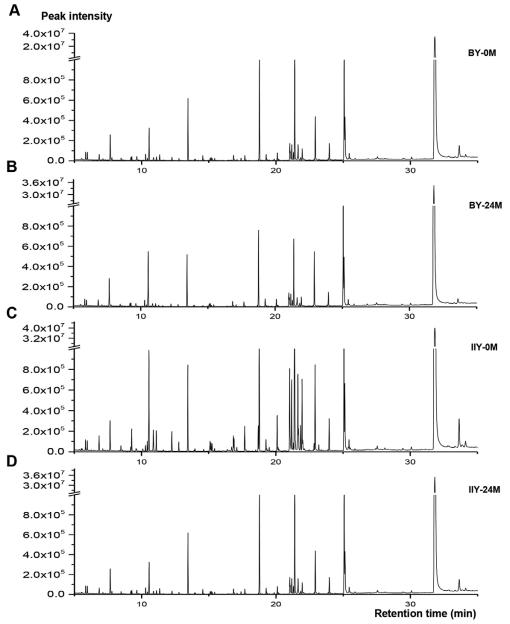 The total ion chromatograms of non-volatile compounds determined by GC-MS in the hybrid rice seeds of BY-0M (A), BY-24M (B), IIY-0M (C), and IIY-24M(D).
The total ion chromatograms of non-volatile compounds determined by GC-MS in the hybrid rice seeds of BY-0M (A), BY-24M (B), IIY-0M (C), and IIY-24M(D).
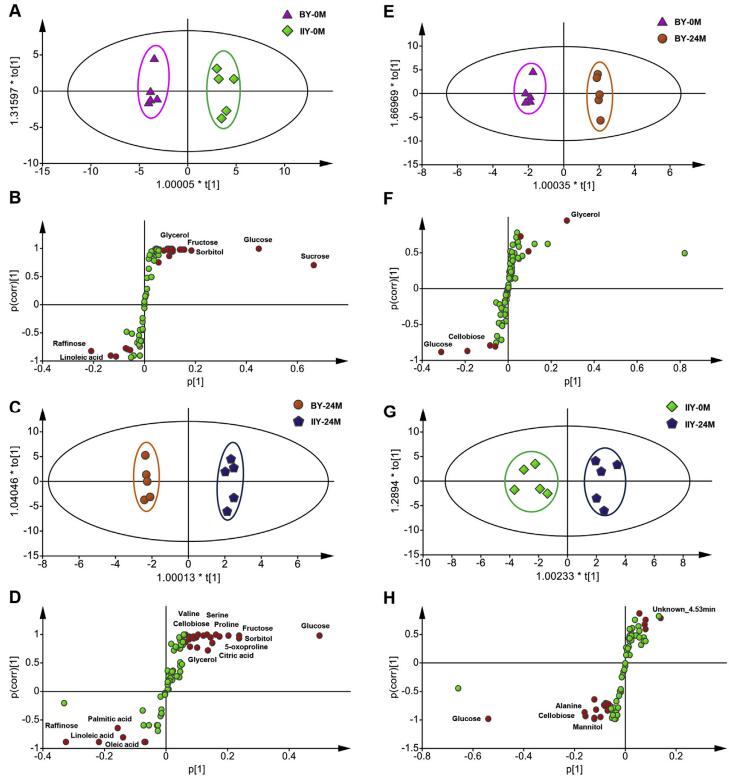 The score plots (A, C, E, G) and S-plot of the different metabolites (B, D, F, H) generated from OPLS-DA of pairwise GC-MS data comparison
The score plots (A, C, E, G) and S-plot of the different metabolites (B, D, F, H) generated from OPLS-DA of pairwise GC-MS data comparison
Reference
- Yan, Shijuan, et al. "Comparative metabolomic analysis of seed metabolites associated with seed storability in rice (Oryza sativa L.) during natural aging." Plant Physiology and Biochemistry 127 (2018): 590-598.



 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service The total ion chromatograms of non-volatile compounds determined by GC-MS in the hybrid rice seeds of BY-0M (A), BY-24M (B), IIY-0M (C), and IIY-24M(D).
The total ion chromatograms of non-volatile compounds determined by GC-MS in the hybrid rice seeds of BY-0M (A), BY-24M (B), IIY-0M (C), and IIY-24M(D). The score plots (A, C, E, G) and S-plot of the different metabolites (B, D, F, H) generated from OPLS-DA of pairwise GC-MS data comparison
The score plots (A, C, E, G) and S-plot of the different metabolites (B, D, F, H) generated from OPLS-DA of pairwise GC-MS data comparison

