What are Plant Primary Metabolites?
Plant primary metabolites are vital organic compounds that play crucial roles in essential metabolic pathways and are indispensable for plant growth, development, and reproduction. These metabolites encompass a range of molecules such as carbohydrates, amino acids, nucleotides, organic acids, and lipids. They are synthesized through diverse biochemical processes and are fundamental for energy production, structural integrity, and intercellular signaling within plants.
The analysis of plant primary metabolites holds immense significance in comprehending plant physiology and biochemistry. It offers valuable insights into the metabolic pathways involved in plant growth, nutrient absorption, stress responses, and defense mechanisms. Through the profiling and quantification of these metabolites, researchers can obtain a comprehensive understanding of the plant's overall metabolic state. Moreover, they can identify metabolic markers associated with specific physiological conditions or environmental factors, paving the way for the development of strategies to enhance plant productivity, stress tolerance, and nutrient utilization.
Plant Primary Metabolite Analysis Services by Creative Proteomics
Creative Proteomics, a leading provider of advanced analytical services, offers a comprehensive range of plant primary metabolite analysis projects. These projects utilize state-of-the-art mass spectrometry-based technologies to identify and quantify primary metabolites in various plant species. Here are some specific projects provided by Creative Proteomics:
Metabolite Profiling: This project aims to comprehensively profile primary metabolites in plant samples, providing a comprehensive overview of the plant's metabolic composition. It involves the identification and quantification of carbohydrates, amino acids, nucleotides, organic acids, and other metabolites using liquid chromatography-mass spectrometry (LC-MS) or gas chromatography-mass spectrometry (GC-MS) techniques.
Targeted Metabolite Analysis: In this project, Creative Proteomics focuses on the analysis of specific primary metabolites or metabolic pathways of interest. By employing LC-MS or GC-MS techniques, targeted metabolite analysis allows for precise quantification and identification of metabolites involved in one-carbon metabolism and related pathways.
Isotope Tracing Studies: Isotope tracing studies provide valuable insights into the metabolic flux and dynamics of primary metabolites within plants. Creative Proteomics offers stable isotope labeling coupled with mass spectrometry analysis to track the incorporation of isotopically labeled compounds into metabolic pathways, enabling the determination of metabolic rates and pathway activities.
Techniques and Instrumentation for Plant Primary Metabolites Analysis
Liquid Chromatography-Mass Spectrometry (LC-MS): Creative Proteomics utilizes high-performance liquid chromatography (HPLC) coupled with high-resolution mass spectrometry for the separation, detection, and quantification of primary metabolites. The LC-MS system used is the Agilent 1290 Infinity II LC System coupled with the Thermo Scientific Orbitrap Exploris 480 Mass Spectrometer. This powerful combination offers excellent sensitivity, selectivity, and allows for the identification and quantification of a wide range of metabolites.
Gas Chromatography-Mass Spectrometry (GC-MS): GC-MS is employed for the analysis of volatile and semi-volatile metabolites. It involves the separation of metabolites using gas chromatography and their subsequent detection and quantification using mass spectrometry. Creative Proteomics employs the Agilent 7890B Gas Chromatograph coupled with the Thermo Scientific ISQ Single Quadrupole Mass Spectrometer for GC-MS analysis. This system ensures reliable separation and detection of metabolites.
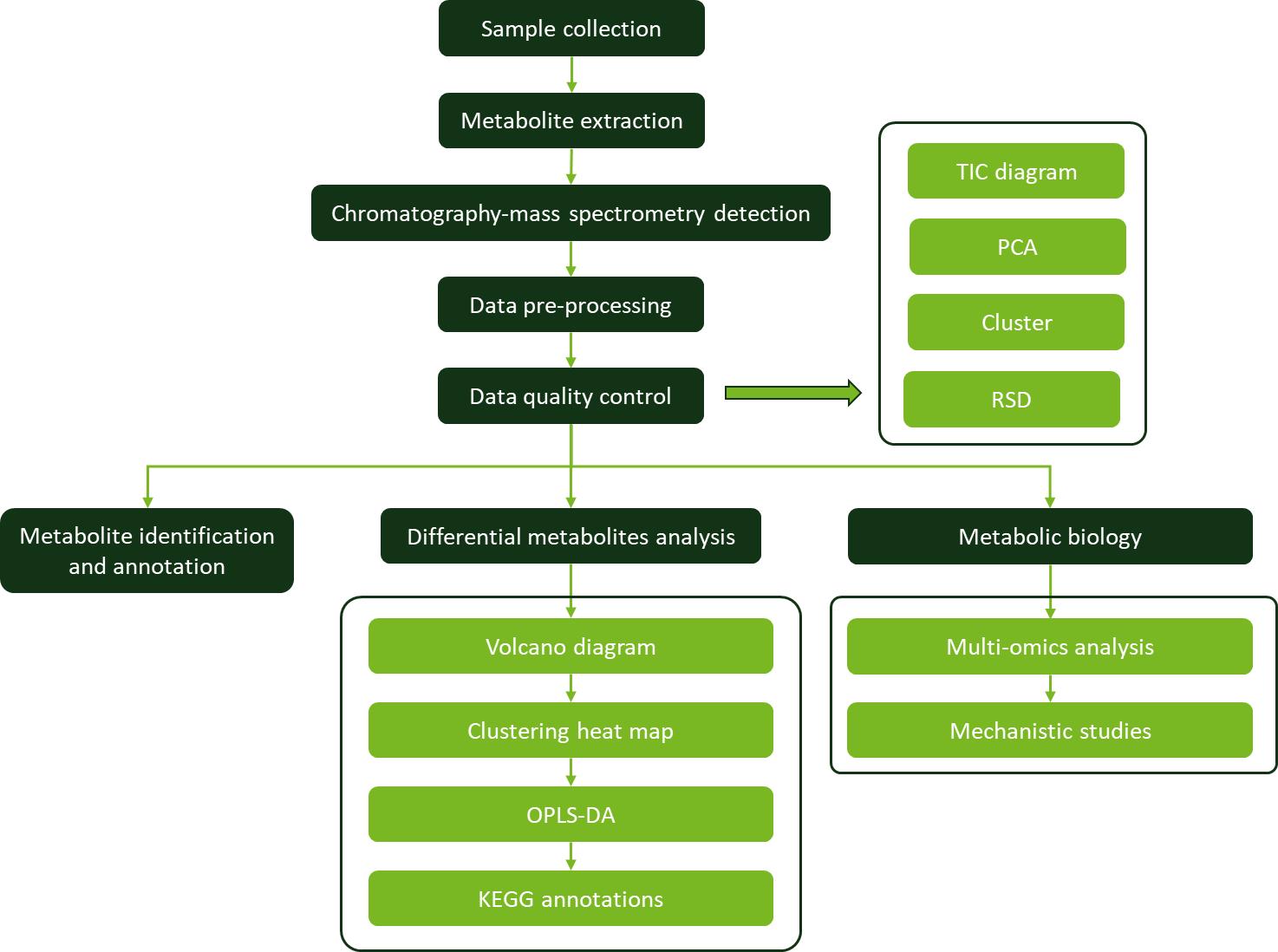 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
List of Plant Primary Metabolites (including but not limited to)
Why Choose Us?
- Detection Platform: Untargeted Metabolomics + Targeted Metabolomics techniques are used to detect metabolites, ensuring both qualitative and quantitative accuracy.
- Comprehensive Database: A self-built plant species-specific metabolite database with over 1800 substances.
- Diverse Range of Metabolites: Primary metabolites such as sugars, amino acids, lipids, nucleotides, organic acids, and more are included.
- Stable Detection: Six major quality control measures are implemented to provide high-quality data.
- Leading Number of Detected Substances: 500-1000 metabolites can be detected in a single analysis.
Applications of Plant Primary Metabolomics
Plant Physiology and Biochemistry: Plant primary metabolite analysis helps elucidate the biochemical processes underlying plant growth, development, and responses to environmental stimuli. It enables the identification of key metabolites involved in energy production, nutrient assimilation, and stress responses.
Crop Improvement and Breeding: By analyzing the metabolic profiles of different plant varieties or genotypes, researchers can identify metabolites associated with desirable traits such as disease resistance, yield potential, and nutritional quality. This information can be used to guide crop improvement and breeding programs.
Stress Physiology and Environmental Adaptation: Metabolomics studies provide insights into how plants respond to abiotic and biotic stresses, such as drought, heat, salinity, and pathogen attacks. By identifying stress-responsive metabolites, researchers can develop strategies for enhancing stress tolerance in crops and promoting sustainable agriculture.
Pharmaceutical and Nutraceutical Research: Plant primary metabolites are a rich source of bioactive compounds with potential pharmaceutical and nutraceutical applications. Metabolomics analysis enables the identification and characterization of these compounds, facilitating drug discovery, natural product research, and the development of functional foods.
Sample Requirements for Plant Primary Metabolites Assay
| Sample Types |
Minimum Sample Size |
Biological Repeat |
| Plant Tissues |
Stem, bud, node, leaf, flower, fruit, root, callus tissue |
600mg |
3-6 |
| Liquid Samples |
Root exudates |
10mL |
| Fermentation broth, wine, tissue fluid, fruit juice |
5mL |
| Honey, nectar, oil, extract |
500μL |
| Specialty Samples |
Cultured samples, presence of liquid |
600mg |
Case 1. Metabolic Analysis Reveals Altered Sulfur Metabolism in astol1 Mutant Rice under Arsenite Tolerance
Background:
Metabolites involved in sulfur metabolism were analyzed.
The analysis aimed to understand the metabolic changes associated with the astol1 mutant and its response to arsenite tolerance.
Samples:
Roots and shoots of 5-week-old wild-type rice (cv. Kasalath) and astol1 heterozygous mutant were used.
Three biological replicates were included for each line, with six plants for each biological replicate.
Technical methods procedure:
Freeze-dried samples were prepared from roots and shoots of the rice plants.
Metabolites were extracted from the samples using 80% ethanol.
Liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) was used for qualitative analysis of metabolites.
Metabolites were quantified using standard curves.
The concentration of sulfate in roots and shoots was determined using an ion chromatography system.
Results
The metabolites involved in sulfur metabolism were analyzed and quantified.
Changes in metabolite levels between wild-type and astol1 mutant were determined.
Specific metabolites and their concentrations were reported as a result of the analysis.
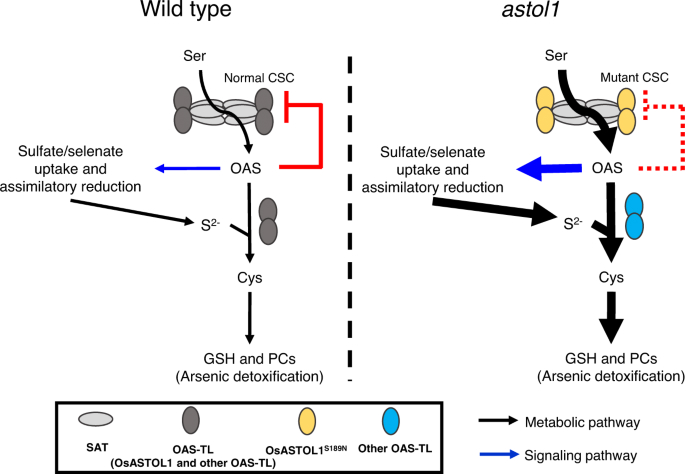 astol1 mutant enhances S and Se uptake and phytochelatins-dependent As detoxification.
astol1 mutant enhances S and Se uptake and phytochelatins-dependent As detoxification.
Reference
- Sun, Sheng-Kai, et al. "A molecular switch in sulfur metabolism to reduce arsenic and enrich selenium in rice grain." Nature Communications 12.1 (2021): 1392.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service astol1 mutant enhances S and Se uptake and phytochelatins-dependent As detoxification.
astol1 mutant enhances S and Se uptake and phytochelatins-dependent As detoxification.

