Lignin, a remarkable biopolymer, constitutes an integral part of plant cell walls. It is a complex and robust phenolic compound resulting from the polymerization of three primary monolignols: p-coumaryl, coniferyl, and sinapyl alcohols. Lignin's intricate and three-dimensional network contributes to the mechanical strength, rigidity, and impermeability of plant tissues. This phenolic marvel is not only pivotal for structural support but also plays a role in defense against pathogens, maintaining water balance, and facilitating nutrient transport within plants.
Creative Proteomics's experts is dedicated to unraveling the mysteries of lignin metabolism. We utilize cutting-edge technologies and advanced methodologies to provide you with accurate and insightful data. Through our analysis, you can gain a deeper understanding of lignin's role in plant growth, stress responses, and applications in various industries. We are committed to helping you uncover the hidden potential within lignin and translating it into practical solutions.
Lignin Detection Projects at Creative Proteomics
Metabolite Profiling: In-depth analysis of lignin-related metabolites using advanced techniques such as LC-MS and GC-MS, enabling comprehensive insights into metabolic pathways.
Metabolic Pathway Mapping: Elucidation of lignin metabolic pathways through targeted and untargeted metabolomics approaches, facilitating a comprehensive understanding of lignin metabolism.
Isotope Tracing Studies: Utilization of isotopic labeling to trace the fate of lignin-derived metabolites, revealing metabolic transformations and fluxes.
Quantitative Metabolomics: Accurate quantification of lignin-associated metabolites to discern concentration changes under different conditions or genetic modifications.
Biomarker Discovery: Identification of potential biomarkers indicative of lignin metabolism alterations, contributing to disease or stress-related studies.
Metabolomics Profiling in Microbial Systems: Exploration of lignin metabolism within microbial communities, aiding the development of lignin-based bioproducts.
Customized Metabolomics Strategies: Tailored experimental designs and data analyses to address specific questions regarding lignin metabolism, ensuring comprehensive support for unique research objectives.
Cutting-Edge Bioinformatics: Utilization of advanced bioinformatics tools for data processing, visualization, and interpretation, maximizing the value extracted from lignin metabolomics datasets.
Technologies Employed for Lignin Analysis
- Liquid Chromatography-Mass Spectrometry (LC-MS): Agilent 1290 Infinity LC System coupled with Agilent 6550 iFunnel Q-TOF Mass Spectrometer, offering precise analysis of lignin metabolites with exceptional sensitivity and resolution.
- Gas Chromatography-Mass Spectrometry (GC-MS): Thermo Scientific TRACE 1310 GC System paired with Thermo Scientific ISQ LT Single Quadrupole Mass Spectrometer, enabling comprehensive profiling of volatile and semi-volatile lignin-derived compounds.
- High-Resolution Mass Spectrometry (HRMS): Waters Xevo G2-XS QTOF Mass Spectrometer, providing accurate mass identification and characterization of lignin-related metabolites with outstanding precision.
- Isotope Ratio Mass Spectrometry (IRMS): Thermo Scientific Delta V Plus IRMS System, facilitating isotopic analysis for tracing lignin metabolites and unveiling metabolic pathways.
- Tandem Mass Spectrometry (MS/MS): AB Sciex TripleTOF 6600 LC-MS/MS System, empowering structural elucidation and fragmentation analysis of compounds associated with lignin metabolism.
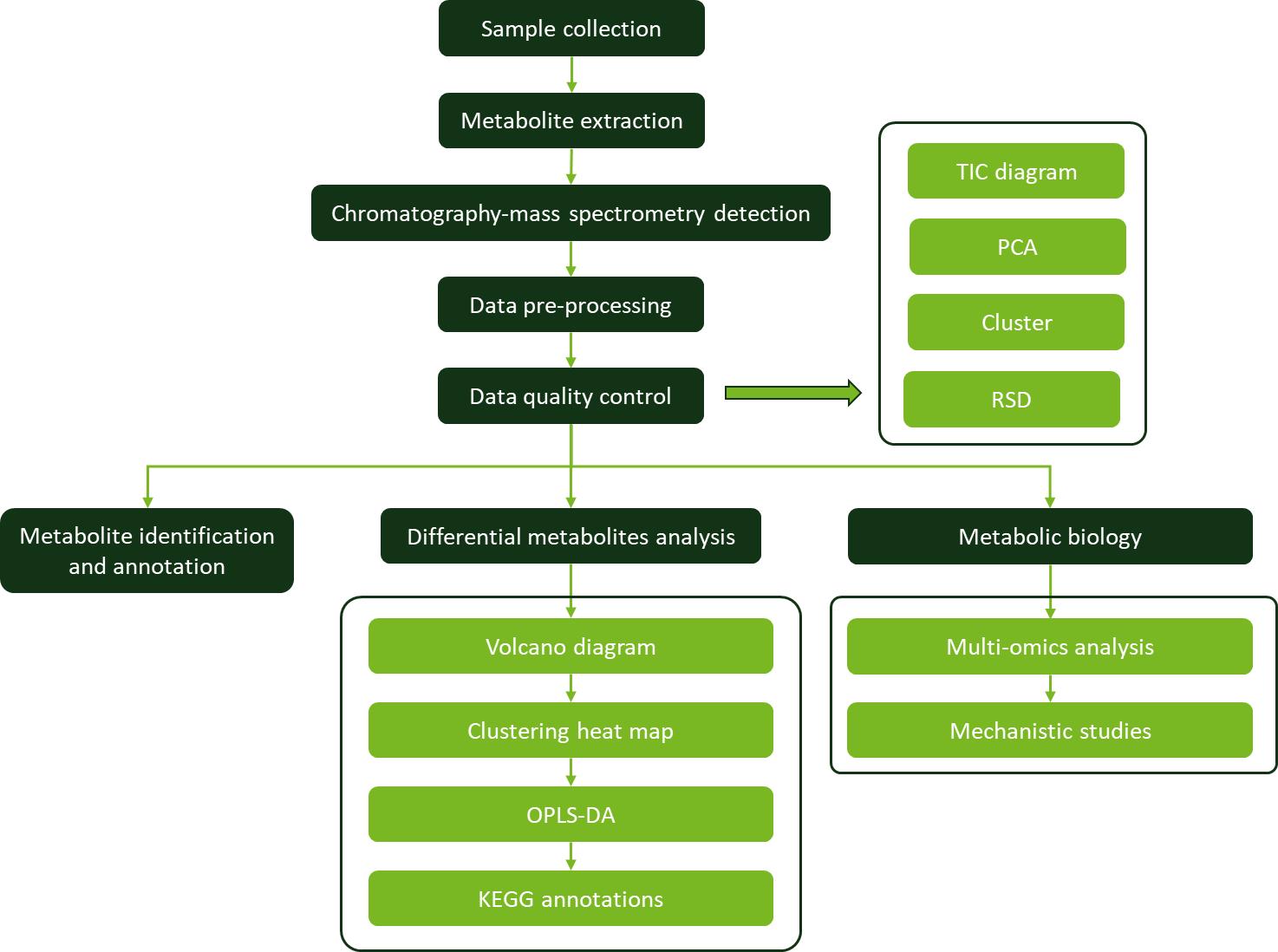 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service
List of Lignin and Metabolites Analyzed (including but not limited to)
| Types |
Lignin and Metabolites |
| Lignin Monomers |
Coniferyl alcohol, Sinapyl alcohol, p-Coumaryl alcohol, Caffeic acid, 5-Hydroxyconiferyl alcohol, 5-Hydroxyconiferaldehyde |
| Lignin Dimers/Trimers |
Dicaffeoylquinic acid, Sinapyl ferulate, Coniferyl β-D-glucoside, Sinapyl β-D-glucoside, Coniferyl sinapate, Sinapyl sinapate, Hexacosyl p-coumaryl ether |
| Lignin Oligomers/Polymers |
Lignosulfonates, Lignocellulosic fibers, Lignin-carbohydrate complexes, Low molecular weight lignin, High molecular weight lignin, Cornified lignocellulose, Hexacosyl p-coumaryl ether |
| Lignin Derivatives |
Vanillin, Syringaldehyde, Ferulic acid, Coumaric acid, Vanillic acid, Syringic acid, Feruloyl glycerol, Coumaroyl glycerol, Vanillic acid glucoside, Syringic acid glucoside, Ferulic acid glucoside, Coumaric acid glucoside |
| Lignin Metabolites |
Vanillic acid, Syringic acid, Feruloyl glycerol, Coumaroyl glycerol, Aryl glycerol-β-guaiacyl ether, p-Hydroxybenzoic acid, Protocatechuic acid, Cinnamic acid derivatives, Lignan metabolites, Phenolic acids, Lignin breakdown products, Cornified lignocellulose metabolites, Hexacosyl p-coumaryl ether derivatives |
Sample Requirements for Lignin Assay
| Sample Types |
Minimum Sample Size |
| Plant Samples |
Roots, stems and leaves, floral parts, fruits/seeds, rhizomes, buds/tender leaves, tissue sections, pollen, bark, trunk/wood, resin/gum, resin acids, seedlings/young plants, rhizosphere soil, root exudates. |
50 mg - 1 g |
Applications of Lignin Metabolomics Analysis
Elucidating Lignin Biosynthesis Pathways: By analyzing lignin metabolites, it reveals the biosynthesis pathways and regulatory mechanisms of lignin. This aids in understanding how plants synthesize different types and amounts of lignin.
Optimizing Bioenergy Production: Lignin is a major component of plant cell walls with implications for bioenergy production. Insights from lignin metabolomics can lead to improved biofuel production methods and enhanced energy production efficiency.
Enhancing Crop Stress Resilience: Lignin plays a vital role in plant responses to stressors like diseases, drought, and salt stress. Lignin metabolomics helps uncover lignin's role in stress responses, contributing to the development of more stress-tolerant crop varieties.
Developing Environmental Adaptation Strategies: Analyzing changes in lignin metabolism under varying environmental conditions informs the breeding of crops adapted to climate change and environmental pressures.
Discovery of Pharmaceuticals and Bioactive Compounds: Lignin and its metabolites may possess bioactivity, serving as potential drug candidates. Lignin metabolomics aids in identifying molecules with therapeutic potential.
Regulation of Plant Growth and Development: Lignin metabolism influences plant growth and development. Studying lignin metabolomics reveals its mechanisms in plant growth regulation.
Environmental Pollution Monitoring: Lignin metabolomics can be used to monitor the degradation and metabolism of organic compounds in the environment, assessing environmental pollution levels.
Plant Evolution and Ecological Studies: Comparing lignin metabolism across different species unveils insights into plant evolution and adaptation to diverse ecological environments.
Case 1. Lignin Degradation Study by Paenibacillus glucanolyticus Strains SLM1 and 5162
Background:
The study delves into the capacity of Paenibacillus glucanolyticus bacteria to degrade lignocellulosic compounds, with a particular emphasis on lignin. Prior research demonstrated the potential of these bacteria to grow on lignin and its components, motivating an in-depth exploration of their lignin degradation capabilities.
Samples:
Two strains of Paenibacillus glucanolyticus, SLM1 and 5162, were selected as subjects for the study. These strains were isolated from distinct sources: SLM1 from black liquor and 5162 representing a type strain. A range of lignocellulosic substrates was employed, encompassing black liquor, cellulose, hemicellulose, switchgrass lignin, and bioChoice lignin (BCL).
Methods:
Lignin Isolation and Fractionation: The process involved the isolation of lignin from switchgrass and subsequent fractionation. This fractionation utilized a combination of acetone and hexanes, resulting in the collection of distinct lignin fractions, each subjected to subsequent analysis.
Lignin Growth Studies: To assess bacterial growth on lignin and lignin-related compounds, optical density (OD) assays were performed. A spectrum of lignin monomers was included in the growth studies to gauge the strains' responses.
Dye Degradation Assays: The study evaluated the bacteria's ability to degrade lignin-like compounds through comprehensive dye degradation assays, encompassing both solid and liquid media.
GC-MS Analysis: Gas chromatography-mass spectrometry (GC-MS) was a pivotal technique employed to scrutinize the production of organic acids during the bacterial growth process on lignin.
GPC Analysis: Gel permeation chromatography (GPC) was applied to analyze the molecular weight variations in lignin prior to and post-bacterial growth.
Results
Both P. glucanolyticus strains, SLM1 and 5162, demonstrated robust growth across a diverse array of lignocellulosic substrates, which included black liquor, cellulose, hemicellulose, and lignin. SLM1 exhibited notably swifter growth kinetics and a higher degree of lignin degradation efficiency in comparison to strain 5162. The dye degradation assays underscored SLM1's proficiency in breaking down lignin-related compounds. GC-MS analysis provided concrete evidence of the production of various organic acids during the lignin degradation process. GPC analysis furnished quantitative insights, indicating a discernible reduction in lignin molecular weight subsequent to bacterial growth, lending support to the notion of lignin depolymerization executed by these strains.
In summation, the study conducts an intricate examination of P. glucanolyticus strains SLM1 and 5162, probing their capacity to degrade lignin and analogous compounds within diverse lignocellulosic substrates. The comprehensive technical arsenal, encompassing lignin isolation, growth studies, dye degradation assays, GC-MS, and GPC analysis, culminates in the identification of these strains as proficient lignin degraders, with SLM1 displaying superior efficiency in this biodegradation process.
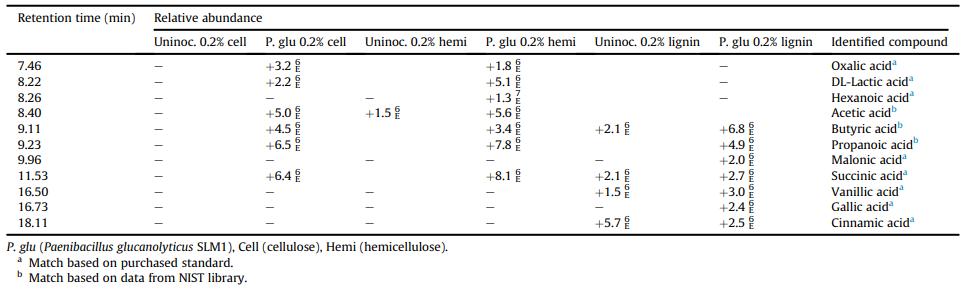 Relative abundance of TMS organic acids identified by GCeMS in Paenibacillus glucanolyticus SLM1 culture supernatant after 400 h of growth in M9 minimal media with the specified carbon source.
Relative abundance of TMS organic acids identified by GCeMS in Paenibacillus glucanolyticus SLM1 culture supernatant after 400 h of growth in M9 minimal media with the specified carbon source.
Reference
- Mathews, Stephanie L., Amy M. Grunden, and Joel Pawlak. "Degradation of lignocellulose and lignin by Paenibacillus glucanolyticus." International Biodeterioration & Biodegradation 110 (2016): 79-86.


 Workflow for Plant Metabolomics Service
Workflow for Plant Metabolomics Service Relative abundance of TMS organic acids identified by GCeMS in Paenibacillus glucanolyticus SLM1 culture supernatant after 400 h of growth in M9 minimal media with the specified carbon source.
Relative abundance of TMS organic acids identified by GCeMS in Paenibacillus glucanolyticus SLM1 culture supernatant after 400 h of growth in M9 minimal media with the specified carbon source.

