Protein ubiquitination, a post-translational modification (PTM) of ubiquitin, has become one of the most active areas of research in biology due to the important role of this PTM in the regulation of various cellular proteins. Creative Proteomics is a leading international PTM proteomics contract research organization (CRO) company. We specialize in applying optimized modified peptide enrichment techniques, advanced mass spectrometry (MS) technologies and novel bioinformatics analysis tools for protein PTM analysis. Here, we offer ubiquitination site identification service by MS-based analysis of tryptic peptide derivatives. The determination and site-specific mapping of protein ubiquitination can help to elucidate the molecular mechanisms of such modifications.
Protein ubiquitination
Ubiquitination serves as a versatile PTM that mediates the growth and development of all eukaryotic species. Ubiquitin (Ub) is a small molecule protein consisting of 76 amino acids, which is characterized as stable, highly conserved, and universally expressed. Compared to protein phosphorylation, the Ub system is vastly more complex. In brief, Ub is conjugated in a specific manner to the target protein or to an already attached Ub chain on the target protein, catalyzed sequentially by the three classes of enzymes, Ub-activating enzyme (E1), Ub-conjugating enzyme (E2), and Ub-ligating enzymes (E3). Among them, E3 determines the specific recognition of target proteins. Ubiquitination is involved in regulating protein degradation, gene transcription, DNA repair and replication, intracellular trafficking, and other fundamental cellular processes. Increasing experimental evidence suggests that changes in the Ub system are closely associated with cellular transformation, inflammatory response, and immune response.
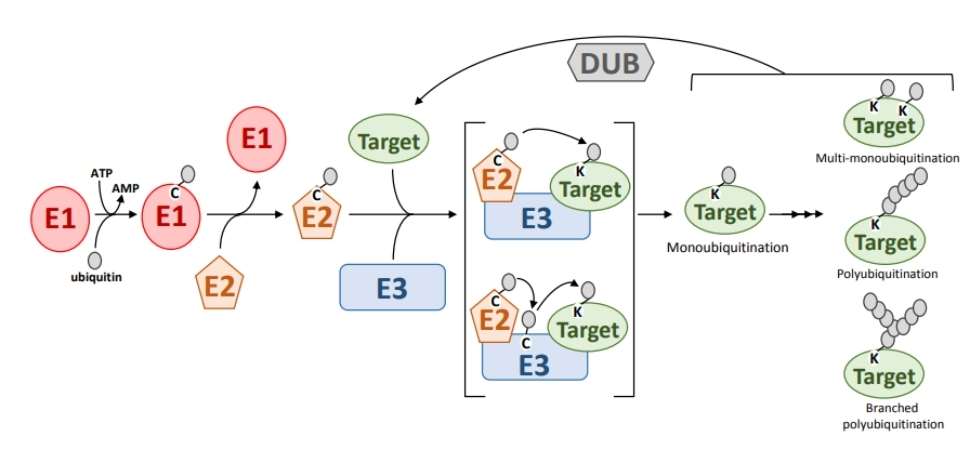 Fig. 1 Ubiquitination pathway showing the function of the ubiquitin enzymes, E1, E2 and E3. (Stone, Sophia L., 2019)
Fig. 1 Ubiquitination pathway showing the function of the ubiquitin enzymes, E1, E2 and E3. (Stone, Sophia L., 2019)
Protein ubiquitination site identification service
Based on our professional platform, we provide detailed and comprehensive services to help clients better solve their research problems. Specifically, our services include experimental design, sample preparation (protein extraction, digestion, peptide, isolation, and enrichment), LC-MS/MS analysis, MS data analysis, and a variety of bioinformatics analysis. Therefore, we can help customers to achieve a large-scale identification of ubiquitinated proteins and ubiquitination sites. In addition, we apply a variety of machine-learning methods to predict protein ubiquitination, which can provide simple and rapid research solutions with valuable information. Our experts can help customers achieve a combination of accurate prediction of ubiquitination sites and validation by ubiquitination MS/MS analysis.
Sample requirements
- Acceptable samples:
-Protein extracts (total protein > 500 mg);
-Cell samples (cell volume > 107);
-Tissue samples (animal tissue >2000 mg, plant tissue >1000 mg);
-Body fluid samples (blood, urine).
-Microorganism dry weight > 200 mg. - Sample shipping: Sufficient amount of dry ice for shipping, or consult our technical staff before sending samples.
- Before the formal experiment, we will always test the samples you provide.
Advantages of our service
- Optimized and upgraded methods and higher depth data detection promote a much higher number of modification sites identified.
- Providing researchers and professionals with a reliable, effective, and end-to-end workflow for ubiquitination site identification.
- Strong R&D team with years of experience in MS, which is steadily leading in the industry.
- Excellent after-sales service.
Creative Proteomics works on a wide variety of PTM analyses to help obtain faster and more accurate molecular mechanisms of these modifications. We are always looking to connect with researchers and professionals who are dedicated to PTM analysis. Here, we are dedicated to helping our customers identify ubiquitination sites from proteins in complex mixtures. Please contact us by leaving a comment, we would love to hear from you.
References
- Udeshi, Namrata D., et al. "Large-scale identification of ubiquitination sites by mass spectrometry." Nature protocols 8.10 (2013): 1950-1960.
- Stone, Sophia L. "Role of the ubiquitin proteasome system in plant response to abiotic stress." International review of cell and molecular biology 343 (2019): 65-110.
Our products and services are for research use only.

 Fig. 1 Ubiquitination pathway showing the function of the ubiquitin enzymes, E1, E2 and E3. (Stone, Sophia L., 2019)
Fig. 1 Ubiquitination pathway showing the function of the ubiquitin enzymes, E1, E2 and E3. (Stone, Sophia L., 2019)