Protein post-translational modifications (PTMs) have key roles in protein folding, activity, function, cellular location, and interactions with other proteins. With the rapid development of proteomics technology, there has been remarkable progress in protein PTM identification and functional analysis. The computational analysis of PTMs is an attractive alternative to traditional experimental methods due to its high accuracy, speed, and low cost. Creative Proteomics is a leading custom service provider in PTM proteomics analysis. Our expert team has many years of experience in the bioinformatics analysis of protein PTMs. We now provide the PTM site prediction service which is part of our integrated portfolio of protein PTM bioinformatics analysis services.
Overview of PTM site identification
The prediction of protein PTMs is currently an important research topic in the field of protein bioinformatics. PTMs allow proteins to adopt more complex structures, have more sophisticated functions, and perform different tasks more precisely. Different types of PTM can occur at different amino acid residues, and multiple modifications may occur at the same amino acid residue. As the data accumulated, by identifying patterns in the acquired data, researchers tried to infer whether a particular locus has been modified or not. The computational prediction has the advantages of high accuracy, high speed, and low cost, and is a suitable and effective alternative screening method to traditional protein PTM analysis. To date, there are a plethora of computational prediction methods developed for protein PTMs. These methods vary depending on the type of prediction algorithm used, enabling the study of the PTM sites of proteins from their sequences, structures, and evolutionary characteristics. Typically, there are four main steps in the computational approach to predicting a particular type of PTM, including data collection, feature extraction, predictor learning, and performance evaluation.
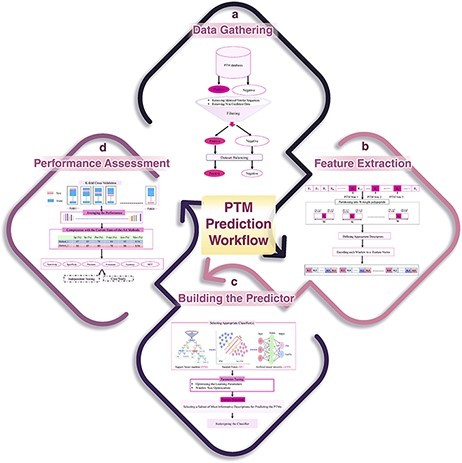 Fig. 1 A schematic flowchart to show how a predictor works for PTM prediction. (Ramazi, S., & Zahiri, J., 2021)
Fig. 1 A schematic flowchart to show how a predictor works for PTM prediction. (Ramazi, S., & Zahiri, J., 2021)
PTM site motif analysis service
Computational prediction of PTMs is a promising strategy to perform preliminary analysis and significantly reduce the number of potential targets requiring further in vivo or in vitro confirmation. Based on our experienced experts and multiple machine learning algorithms, we are able to provide not only common PTM site prediction services, such as phosphorylation site, glycosylation site, acetylation site, methylation site, SUMOylation site and ubiquitination site prediction, but also new PTM site prediction services. In addition, we are able to predict whether the PTM site is within the protein-protein interaction region (PPIR). PPIR forms the binding interface between interacting polypeptide chains and is a very important functional domain of proteins. PTMs in PPIR can interfere with or promote protein interactions, and their study can help further elucidate the regulatory mechanisms by which PTMs of targeted proteins regulate their cellular functions.
Advantages of our service
- Use more carefully selected and complete PTM-related databases and resources.
- Focus on developing more rigorous computational models and designing more efficient algorithms to enhance PTM research.
- More efficient identification of new PTM substrates on a large-scale level.
In the last several decades, a large amount of protein sequence data has been generated, highlighting the importance of the in-depth study and analysis of protein PTMs. To simplify the analysis of complex PTM data and to improve our clients' understanding of various PTMs in different organisms, Creative Proteomics offers PTM bioinformatics analysis services. Our bioinformatics team has done a lot of work on the identification of PTM results and has developed our own platform for quality control, data processing, and analysis. Contact us for all the details!
Related services
References
- He, Wenying, Leyi Wei, and Quan Zou. "Research progress in protein posttranslational modification site prediction." Briefings in functional genomics 18.4 (2019): 220-229.
- Wang, Duolin, et al. "MusiteDeep: a deep-learning based webserver for protein post-translational modification site prediction and visualization." Nucleic Acids Research 48.W1 (2020): W140-W146.
- Ramazi, Shahin, and Javad Zahiri. "Post-translational modifications in proteins: resources, tools and prediction methods." Database 2021 (2021).
Our products and services are for research use only.

 Fig. 1 A schematic flowchart to show how a predictor works for PTM prediction. (Ramazi, S., & Zahiri, J., 2021)
Fig. 1 A schematic flowchart to show how a predictor works for PTM prediction. (Ramazi, S., & Zahiri, J., 2021)