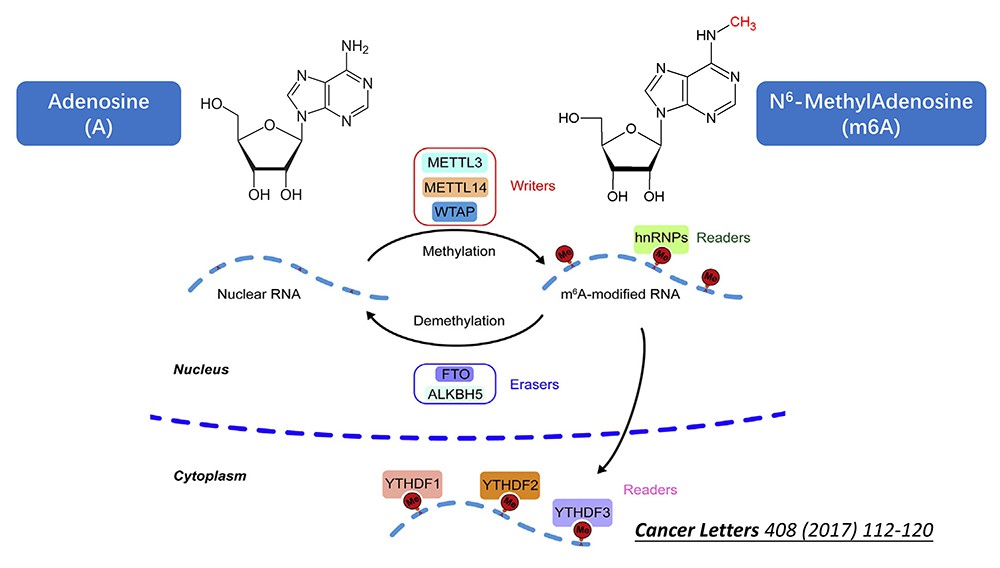
What is m6A Modification
In eukaryotes, there are more than 100 known modifications of RNA, including m6A, m1A, m6Am, m5C, hm5C, etc. m6A is the most common internal modification of eukaryotic mRNA, accounting for 80% of RNA methylation modifications. In humans and mice, 1/3~1/2 of the mRNA has m6A modification, and Arabidopsis has about 1000 bases. 0.5-0.7 m6A peak. Previous studies mainly focused on tRNA and other modifications, and now it is found that there are a large number of m6A modifications inside mRNA, which indicates that m6A will participate in a variety of physiological and disease functions, but the current mechanism is not well understood, so there is a lot of research value. m6A refers to the methylation modification on the 6th N of RNA adenylate (N6-adenylate methylation), which is a dynamic and reversible modification method.
The biological functions of m6A include: promoting the formation of precursor miRNA, mediating alternative splicing of pre-mRNA, combining IF and ribosomes to affect translation efficiency, driving translation initiation, mediating mRNA degradation, and exercising various functions in the nucleus in conjunction with other reader proteins. function etc. In tumor research reports, m6A modification is involved in the tumorigenesis and metastasis of various malignant tumors by regulating RNA splicing, processing, translation, and degradation. However, the underlying mechanism of m6A modification in cancer remains to be studied. And m6A is also increasingly used in other animal and even plant research.
 Quantitative detection of m6A by mass spectrometry
Quantitative detection of m6A by mass spectrometry
The use of mass spectrometry allows for both relative and absolute quantification of m6A levels in multiple samples. Liquid chromatography-mass spectrometry (LC-MS) is a powerful analytical technique that combines the separation capabilities of liquid chromatography with the accurate mass measurement of mass spectrometry. This technique enables the identification and quantification of m6A residues in RNA molecules.
LC-MS/MS, based on liquid chromatography-mass spectrometry, employs tandem mass spectrometry to obtain molecular ion peaks and fragment ion peaks, enabling qualitative and quantitative analysis of nucleotides simultaneously.
As shown in Figure 1, the first step involves the extraction of Total RNA using TRIzol. Following this, mRNA can be enriched using oligo(dT) magnetic beads, or rRNA removal kits can be utilized to obtain various RNA types including mRNA and lncRNA. In the second step, Nuclease P1 is used to digest single-stranded RNA into individual nucleotides. Subsequently, alkaline phosphatase and ammonium bicarbonate are added, and after several hours of incubation, the sample is injected into a liquid chromatography system. The content of each nucleotide is calculated based on the peak area of their retention time. In the fourth step, the sample undergoes tandem mass spectrometry analysis, where individual ribonucleotides are ionized and fragmented into pentose and pyrimidine or purine moieties. Finally, the area of m6A is calculated based on its retention time. The overall methylation level of m6A on mRNA can be determined by calculating the ratio of m6A to total adenine content.
Significance and Applications of Quantitative m6A Detection
Understanding Functional Significance
The quantitative detection of m6A modifications using LC-MS has provided valuable insights into their functional significance in different biological contexts. Researchers have used this method to measure m6A levels in various cell types, developmental stages, and environmental cues to uncover the dynamic nature of m6A regulation. Support from Creative Proteomics has enabled research into the functional implications of m6A modifications.
Revealing RNA Epigenetics
m6A modification plays a critical role in the emerging field of RNA epigenetics. The high-precision quantification of m6A levels can help researchers uncover the epigenetic landscape of RNA molecules and identify novel regulatory mechanisms. Creative Proteomics is a leading provider of cutting-edge technologies and expertise in RNA epigenetics research.
Clinical Applications
Dysregulation of m6A modification has been linked to various diseases, including cancer, neurological disorders, and metabolic disorders. The quantitative detection of m6A by mass spectrometry presents a potential avenue for identifying biomarkers and therapeutic targets.
Our Services
We can provide a complete range of experiments, including total RNA extraction from samples, mRNA enrichment and enzymatic digestion, enzymatic hydrolysis of nucleosides for MRM testing, and statistical analysis of quantitative data. Additionally, we also offer MRM detection services for other types of RNA modifications, as well as related fields such as carbohydrate metabolism, lipid metabolism, and the TCA cycle.
m6A Quantitative Analysis Service Highlights
- State-of-the-art mass spectrometry technology for precise and sensitive detection of m6A modifications.
- Robust sample preparation protocols for efficient and accurate conversion of m6A into a detectable form.
- Quality-controlled calibration standards for accurate quantification of m6A levels.
- Customized data analysis and interpretation by expert bioinformaticians and data scientists.
- Collaborative approach and expert support throughout the entire process.
- Strict confidentiality and data security measures.
- Comprehensive solution for exploring RNA epigenetics and gene expression regulation.
References
- Nature Communications 2019, 10, 1898.
- Cell Stem Cell 2018, 22, 191-205.e199.
Our products and services are for research use only.

 Quantitative detection of m6A by mass spectrometry
Quantitative detection of m6A by mass spectrometry