Hydroxylation, a pivotal biochemical process, involves the enzymatic addition of a hydroxyl group (-OH) to organic compounds. This transformative modification occurs across a spectrum of biomolecules, playing a fundamental role in various physiological and pathological processes. Understanding the nuances of hydroxylation is paramount, given its profound implications in fields ranging from drug development to environmental science.
In the realm of proteins, hydroxylation plays a pivotal role in post-translational modification. A prime example is the hydroxylation of proline and lysine residues in collagen, the most abundant protein in mammals. This modification imparts stability to the collagen triple helix, crucial for the structural integrity of connective tissues such as skin, bone, and cartilage. The precision of hydroxylation events is essential for the proper folding and function of numerous proteins, impacting cellular processes like signal transduction and gene expression.
Analytical Methods of Hydroxylation
Precise analysis of hydroxylation events demands sophisticated analytical techniques that can discern subtle molecular changes. Several cutting-edge methods have been developed to unravel the intricacies of hydroxylated biomolecules.
- Mass Spectrometry. Mass spectrometry stands at the forefront of hydroxylation analysis. High-resolution mass spectrometry enables the identification and quantification of hydroxylated residues within proteins and other biomolecules. This technique provides invaluable insights into the extent and site-specific nature of hydroxylation, empowering researchers to map the molecular landscape with unparalleled precision.
- Nuclear Magnetic Resonance (NMR) Spectroscopy. NMR spectroscopy emerges as a powerful tool for elucidating the structural consequences of hydroxylation. By examining changes in chemical shifts and coupling constants, NMR offers a window into the three-dimensional conformation of hydroxylated molecules, facilitating a comprehensive understanding of the modification's impact on molecular architecture.
- Chromatographic Techniques. Chromatographic methods, including high-performance liquid chromatography (HPLC) and gas chromatography (GC), are instrumental in separating and quantifying hydroxylated compounds. These techniques are particularly valuable in studying hydroxylated metabolites, providing quantitative data that contribute to unraveling metabolic pathways influenced by hydroxylation.
Applications of Hydroxylation
The far-reaching implications of hydroxylation extend across various scientific domains, prompting innovative applications that harness its biochemical versatility.
- Drug Development. In the realm of drug discovery, hydroxylation plays a pivotal role in the metabolism of pharmaceutical compounds. Understanding and manipulating hydroxylation pathways are critical for optimizing drug efficacy and minimizing potential adverse effects. Researchers leverage analytical insights into hydroxylation to tailor drug structures for enhanced bioavailability and metabolic stability.
- Environmental Monitoring. Hydroxylation is intricately linked to the degradation of environmental pollutants. Monitoring hydroxylation events in microbial communities facilitates the assessment of their ability to metabolize and detoxify pollutants. This knowledge is pivotal for designing bioremediation strategies that capitalize on nature's enzymatic prowess to mitigate environmental contamination.
- Disease Biomarkers. Hydroxylation patterns in biomolecules serve as potential biomarkers for various diseases, including cancer and neurodegenerative disorders. The identification of hydroxylation-specific signatures in biofluids opens new avenues for non-invasive diagnostic approaches, offering a glimpse into the molecular alterations associated with pathological conditions.
Our Service Workflow
In the realm of biological sciences, hydroxylation stands as a dynamic and multifaceted process with far-reaching implications. From its structural impact on biomolecules to its applications in drug development and environmental science, the study of hydroxylation continues to unveil new dimensions in cellular regulation and adaptation. Creative Proteomics relies on advanced analysis techniques, continue to dissect the complexities of hydroxylation expands, opening avenues for groundbreaking discoveries and innovative applications in diverse scientific disciplines. Creative Proteomics is dedicated to the field of protein research and we provide comprehensive, reliable protein modification analysis services. If you are interested in our services, please feel free to contact us.
Widespread hydroxylation of unstructured lysine-rich protein domains by JMJD6
Journal: PNAS
Published: 2022
Background
Hydroxylation on proteins is an important post-translational modification. Hydroxylation of proline on the classical hypoxia-inducible factor HIF can help cells to sense hypoxic conditions and initiate downstream transcription. In addition to proline, hydroxylation on lysine, aspartate, histidine and other residues has also been reported in recent years, and the enzymes that catalyze the generation of this modification include not only α-ketoglutarate-dependent dioxygenases, but also JMJD family proteins, among which JMJD6 has been reported to be involved in a variety of biologically important processes, but the exact substrate and molecular mechanism are still unknown.
Results
In order to discover substrates for hydroxylation modifications from the proteome, the authors identified one/two hydroxylation modifications at positions 520 to 537 of the BRD4 protein from 12.6 million spectra by depth-graded proteomics.It has been reported in the literature in 2019 that the proline at position 536 of BRD4 can be hydroxylated and modified by the catalytic action of PHD2 protein, but the authors' library search results gave a scoring results that tend to suggest that the modification occurs at position 537. Repeating previous experiments, the authors found that knockdown of the PHD1-3 proteins had no effect on the level of identified hydroxylation modifications, whereas the hydroxylation modifications completely disappeared after knockdown of the known BRD4-interacting protein, JMJD6, suggesting that JMJD6 is the enzyme that actually catalyzes the hydroxylation modifications on BRD4 (Figure 1).
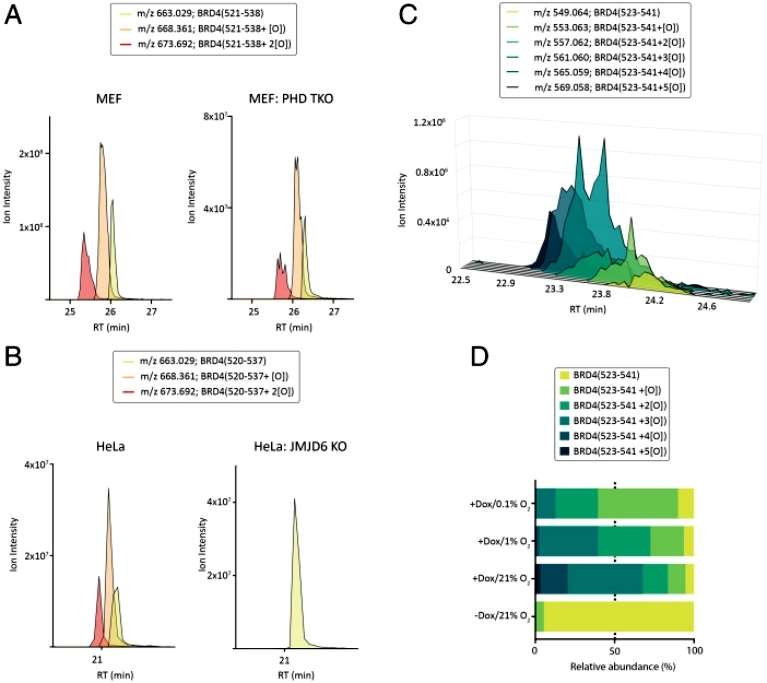 Figure 1
Figure 1
Subsequently, in order to determine at which site the hydroxylation modification occurred, and considering that the C-terminal end of BRD4 is lysine-rich and that the traditional trypsin cleavage could not yield peptides suitable for mass spectrometry, the authors found that switching to Asp-N cleavage yielded peptides of the appropriate lengths and identified five hydroxylation modification sites on BRD4. In order to further improve the coverage of the lysine-rich region, the authors optimized the process to enrich by the ligand of BRD4, and then derivatize the lysine by propionic anhydride to neutralize the positive charge and cause leaky cuts of trypsin to obtain peptides of appropriate length. Ultimately the authors succeeded in identifying 19 hydroxylation modification sites on BRD4, 14 of which were located in the lysine-rich region. Analyzing the modification rates of hydroxylation modifications on these sites they found that some of them were close to 100% (Figure 2).
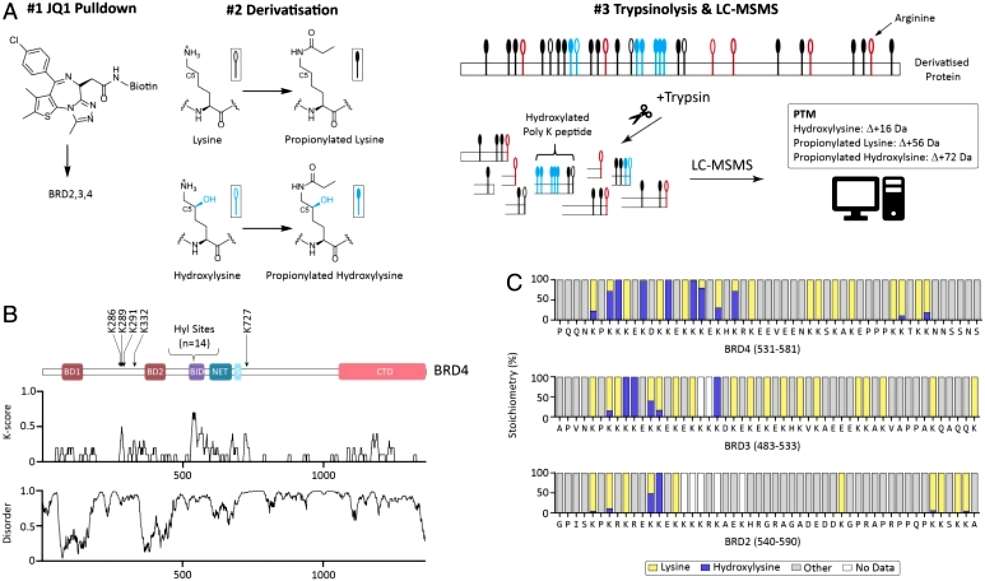 Figure 2
Figure 2
Considering the large number of lysine hydroxylation modifications identified by the authors in the Lys-rich structural domain of BRD4, the authors then hoped to identify more JMJD6 substrates in the proteome, and they augmented JMJD6 binding to the substrate by dimethylglycolylaminoacetic acid (DMOG) as an analog of α-ketoglutaric acid, which in turn pulldown the JMJD6 substrate proteins, and they identified 108 hydroxylation modification sites on 32 proteins by mass spectrometry. On the other hand, they utilized the peptide of the catalytic region of JMJD6 to pull-down directly, and identified a total of 39 modification sites on 26 proteins, and similarly, some of the modification sites had very high modification rates (Figure 3).
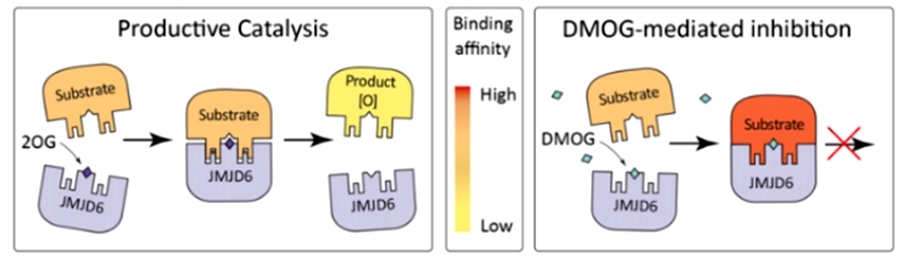 Figure 3
Figure 3
Conclusion
In summary, this article found that JMJD6 catalyzes lysine hydroxylation modifications on lysine-rich regions of proteins and identified a large number of modification substrates.
Reference
- Cockman ME, Sugimoto Y, Pegg HB, Masson N, Salah E, Tumber A, Flynn HR, Kirkpatrick JM, Schofield CJ, Ratcliffe PJ. Widespread hydroxylation of unstructured lysine-rich protein domains by JMJD6. Proc Natl Acad Sci U S A. 2022 Aug 9;119(32):e2201483119.


 Figure 1
Figure 1 Figure 2
Figure 2 Figure 3
Figure 3