Understanding the intricacies of bacterial glycosylation is a critical frontier in the field of proteomics. Glycosylation, a post-translational modification (PTM), plays a pivotal role in modulating the structure and function of proteins. While eukaryotic glycosylation has been extensively studied, unraveling the complexities of bacterial glycosylation poses unique challenges.
Getting Bacteria to Glycosylate Analysis in PTMs
- Decoding Bacterial Glycosylation Patterns. Bacterial glycosylation involves the enzymatic addition of glycan moieties to proteins, impacting their stability, localization, and interaction with other biomolecules. Analyzing these glycosylation patterns is crucial for deciphering the molecular intricacies of bacterial physiology and pathogenesis.
- Unveiling PTMs in Bacterial Proteins. Post-translational modifications in bacterial proteins, particularly glycosylation, play a pivotal role in regulating cellular processes. This analysis provides a window into the dynamic landscape of PTMs, shedding light on how bacteria modulate their proteins to adapt to various environmental conditions.
Analytical Methods of Getting Bacteria to Glycosylate
- Mass Spectrometry. Mass spectrometry emerges as the cornerstone of getting bacteria to glycosylate Analysis. High-resolution mass spectrometry allows for the accurate identification and quantification of glycosylated peptides, providing a comprehensive view of the glycoproteome.
- Glycan Mapping Techniques. Advanced glycan mapping techniques, such as liquid chromatography and capillary electrophoresis, enable the elucidation of glycan structures attached to bacterial proteins. These methods contribute to building a detailed catalog of bacterial glycosylation patterns.
- Bioinformatics Approaches. In silico analysis, supported by bioinformatics tools, facilitates the interpretation of complex glycoproteomic data. Integration of mass spectrometry results with bioinformatics enhances the accuracy of glycosylation site identification and aids in deciphering the underlying regulatory mechanisms.
Applications of Getting Bacteria to Glycosylate Analysis
- Targeting Antibiotic Resistance Mechanisms. Understanding bacterial glycosylation provides insights into antibiotic resistance mechanisms. By analyzing glycosylation patterns in resistant strains, researchers can identify potential targets for novel antibacterial agents, advancing the fight against antibiotic-resistant infections.
- Vaccine Development. Bacterial glycoproteins often serve as virulence factors and play a crucial role in host-pathogen interactions. Getting Bacteria to Glycosylate Analysis contributes to the development of effective vaccines by elucidating glycosylation patterns associated with pathogenicity.
Significance of Getting Bacteria to Glycosylate Analysis
- Precision Medicine Opportunities. The detailed analysis of bacterial glycosylation opens avenues for precision medicine approaches. By understanding the glycoproteomic landscape, tailored therapeutic interventions can be designed to target specific bacterial strains, minimizing collateral damage to beneficial microbiota.
- Unraveling Host-Microbe Interactions. Getting bacteria to glycosylate analysis is instrumental in unraveling the complex interplay between bacteria and their hosts. By decoding how bacterial glycosylation influences host responses, researchers gain valuable insights into the mechanisms of infection, inflammation, and immune evasion.
Our Service Workflow
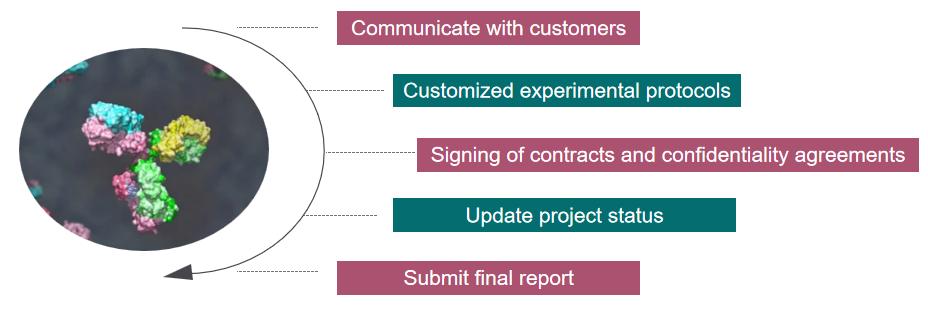
Getting bacteria to glycosylate analysis stands at the forefront of modern proteomics, offering a key to deciphering the intricate language of bacterial glycosylation. From precision medicine to vaccine development, the applications of this analysis are far-reaching, promising a deeper understanding of bacterial biology and novel strategies for combating infectious diseases. Creative Proteomics has rich experience in glycosylate research, if you are interested in us, please feel free to contact us, we look forward to working with you.
How Sweet Are Our Gut Beneficial Bacteria? A Focus on Protein Glycosylation in Lactobacillus
Journal: Int J Mol Sci
Published: 2018
Background
Protein glycosylation, the attachment of carbohydrate moieties to proteins, is a widespread post-translational modification found in all three domains of life—eukaryotes, archaea, and bacteria. Initially thought to be exclusive to eukaryotic and archaeal systems, recent evidence indicates that glycosylation is a common feature in prokaryotes as well. Approximately 70% of eukaryotic and 50% of prokaryotic proteins undergo glycosylation. In eukaryotes, glycoproteins are modified through N-glycosylation on Asp or O-glycosylation on Ser/Thr, involving pre-assembled N-glycans or direct synthesis of O-glycans. Bacterial glycosylation, however, is more diverse, encompassing various mechanisms and carbohydrate structures. Unlike eukaryotic glycosylation, which can occur co- and post-translationally, prokaryotic glycosylation is predominantly post-translational.
Results
The first fully characterized glycosylation system in bacteria is found in Campylobacter jejuni. This system, known as the protein glycosylation cluster (pgl), consists of 13 genes responsible for glycosylating various proteins. The genes within this cluster encode enzymes for bacilosamine synthesis, glycosyltransferases (GTs) involved in glycan production, a transporter (PglK) for glycan translocation to the periplasm, and an oligosaccharyl-transferase (PglB) responsible for glycosylating the target protein. The glycan structure involves α-GalNAc residues and a diNAcBac (2,4-diacetamido-2,4,6-trideoxyglucose) unit at the reducing end (Figure 1).
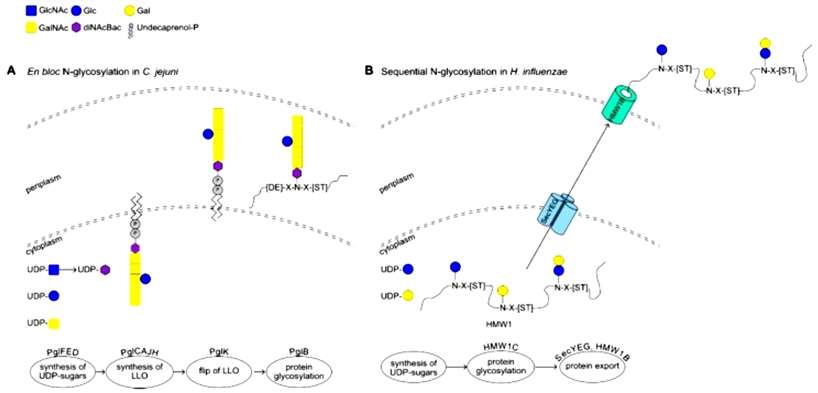 Figure 1
Figure 1
Bacteria, akin to eukaryotes, employ mechanisms for O-glycosylation, modifying proteins on Ser or Thr residues with glycans. Two identified mechanisms for prokaryotic N-glycosylation are: (i) en bloc transfer of a pre-assembled lipid-linked oligosaccharide, and (ii) direct modification of the acceptor protein through the sequential action of glycosyltransferases (GTs). In the en bloc glycosylation mechanism, the glycan is synthesized on undecaprenol phosphate, transferred to the periplasm, and then attached to the acceptor protein by an O-oligosaccharyltransferase (O-OTase). Sequential O-glycosylation involves multiple GTs directly acting on the acceptor protein, using sugar nucleotides as donors to extend the glycan (Figure 2).
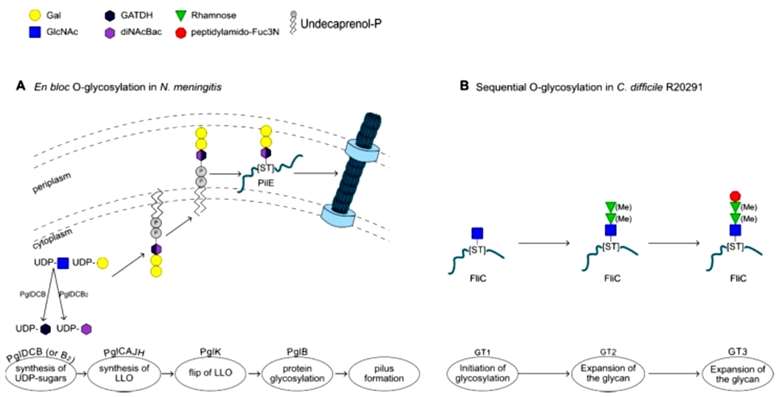 Figure 2
Figure 2
Pathogenic Gram-positive bacteria often possess an auxiliary secretion system called SecA2, alongside the canonical SecA. This system is responsible for the expression, glycosylation, and subsequent secretion of serine-rich repeat (SRR) containing proteins (SRRPs). Streptococcus parasanguinis has a well-studied SecA2 system with unique features not found in other glycosylation systems. The S. parasanguinis SecA2 cluster includes six glycosyltransferases (GTs). The initiation of glycosylation on Fap1, the SRRP in S. parasanguinis FW213, involves two GTs, GtfA and GtfB (or Gtf1 and Gtf2 in some studies). GtfA acts as a GT, while GtfB serves as a chaperone, and both interact with the acceptor SRRP through a conserved domain (DUF1975), mediating the addition of the reducing GlcNAc. Subsequent GTs, such as GtfC, GalT2, dGT1, and Gly, extend the glycan by adding various sugar residues, creating a branching structure in the glycan (Figure 3).
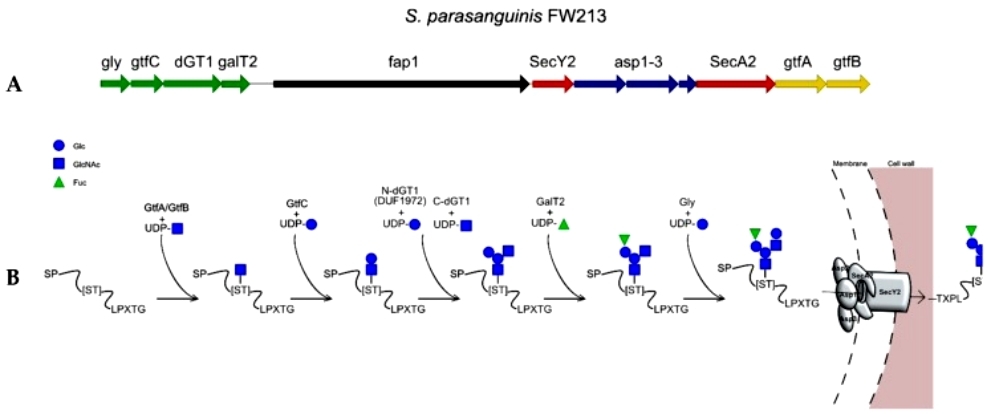 Figure 3
Figure 3
Muramidases, particularly Acm2 in Lactobacillus plantarum strain WCFS1, represent well-characterized glycoproteins in Lactobacillus species. Acm2 is a modular protein with a catalytic domain, an O-glycosylated N-terminal region rich in Ala, Ser, and Thr (AST domain) of unknown function, and a C-terminal region composed of five SH3b peptidoglycan binding domains. Mass spectrometry analysis revealed that Acm2 is glycosylated by single N-acetylhexoseamine (HexNAc) residues at over 20 glycosylation sites within the AST domain. Intriguingly, glycosylation occurs intracellularly before secretion, as demonstrated by the deletion of the secretion signal peptide. The glycosylation of Acm2 was found to partially inhibit its enzymatic activity, possibly through the interaction of N-Acetylglucosamine (GlcNAc) moieties with the active site or SH3b motifs responsible for binding the GlcNAc-rich peptidoglycan layer. Additionally, glycosylation increased the resistance of the AST domain against trypsin (Figure 4).
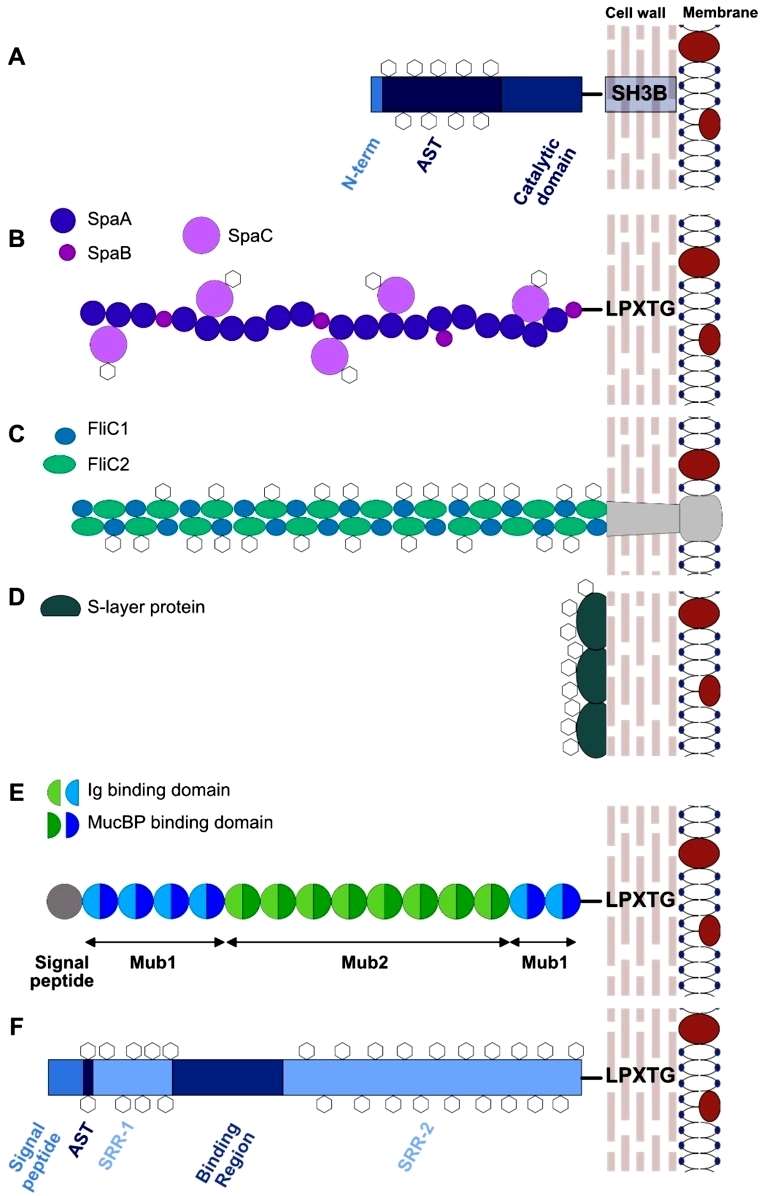 Figure 4
Figure 4
Conclusion
Host-microbe interactions in the gut play a crucial role in determining the outcome of pathogenic infection or commensal colonization, essential for maintaining gut homeostasis. While the significance of protein glycosylation is well-recognized in pathogens, there is a need for increased efforts to comprehend glycosylation in resident gut bacteria. Glycoproteins, in particular, are a relatively understudied yet potentially vital factor influencing bacteria-host interactions, including adhesion, biofilm formation, and immune response. Glycosylation contributes to protein diversity and structure, significantly impacting the function of resulting glycoproteins, potentially influencing bacterial abilities to co-aggregate or auto-aggregate. Recent advancements in structural and analytic tools, such as glycomics, are poised to facilitate further exploration into novel bacterial glycan structures and the characterization of glycosylation pathways in major gut commensal bacteria. This knowledge is crucial for understanding the role of protein glycosylation in shaping bacteria-host interactions and for leveraging the diverse array of bacterial molecules in developing novel therapeutic approaches, including drugs and biomarkers, that target the microbiome.
Reference
- Latousakis D, Juge N. How Sweet Are Our Gut Beneficial Bacteria? A Focus on Protein Glycosylation in Lactobacillus. Int J Mol Sci. 2018 Jan 3;19(1):136.


 Figure 1
Figure 1 Figure 2
Figure 2 Figure 3
Figure 3 Figure 4
Figure 4