Protein S-nitrosylation involves covalently linking nitric oxide (NO) molecule to the thiol group of a cysteine
residue, leading to the formation of S-nitrosothiols that mediate redox-based signaling and primarily convey the
general effects of NO on cellular function. Based primarily on liquid chromatography-tandem mass spectrometry
(LC-MS/MS), Creative Proteomics can help researchers detect the degree of S-nitrosylation of
proteins in cells and tissues under different physiopathological conditions and provide better results.
Introduction of protein S-nitrosylation
S-nitrosylation is a reversible covalent chemical reaction in which a NO is coupled to a critical cysteine thiolate
(-S−) on a target protein, and finally generates an S-nitrosothiol. S-nitrosylation is achieved by triggering
conformational changes in the protein that affect its activity, subcellular localization and interaction with
partners. To date, over 3000 protein candidates have been identified that may undergo S-nitrosylation under normal
and/or pathological conditions. Approaches such as the biotin-switch technique and MS-based proteomics have
contributed greatly. S-nitrosylated proteins cover a wide range of cellular functions, such as receptors, metabolic
enzymes, ion channels, signaling proteins, proteases, and structural proteins. In addition, the removal of NO from
cysteine residues, i.e. denitrosylation, has been found to be a key mechanism for regulating protein activity,
protein-protein interactions, and more generally signal transduction.
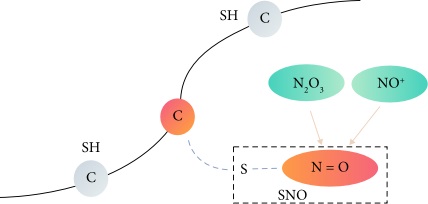 Fig. 1 A schematic
diagram of protein S-nitrosylation sites. (Zhao, Qian, et al., 2021)
Fig. 1 A schematic
diagram of protein S-nitrosylation sites. (Zhao, Qian, et al., 2021)
Proteomics analysis of S-nitrosylation
Increasingly, studies have focused on systematically analyzing the extent of protein S-nitrosylation in tissues and
cells. We have established a highly sensitive HPLC-MS/MS pipeline that can help researchers better understand the
mechanisms by which protein S-nitrosylation occurs. Typically, modified protein samples can be separated using
SDS-PAGE, digested with sequence-specific proteases, and then analyzed in a mass spectrometer connected to a
matrix-assisted laser desorption ionization (MALDI) or electrospray ionization (ESI) ionization source. Peptides
obtained by enzymatic digestion can be further separated by liquid chromatography for easy identification. Relying
on the mass-to-charge shift of a specific PTM, MS can be successfully used to detect and identify S-nitrosylation
modifications. Our optimized method enables faster and more sensitive analysis of S-nitrosylated proteins.
Workflow of our service

Major benefits of our service
- Sample preparation techniques enrich S-nitrosylated proteins to improve detection sensitivity.
- Powerful mass spectrometric analysis.
- Provide detailed reports, including relevant MS parameters, raw data, details of identified glycosylation sites,
mass spectrometric images, etc.
- Provide a variety of bioinformatics analyses.
- One-stop service: Sample processing - Mass spectrometric analysis - Bioinformatics analysis - Project report.
As one of the important PTM of protein, S-nitrosylation regulates multiple biological processes by modulating protein
activity, stability, subcellular localization, and protein-protein interactions. Exploration of the role of
S-nitrosylation in human diseases can assist to clarify the pathogenesis and therapeutic strategies. Our customer
service representatives are available 24 hours a day, 7 days a week. Please feel free to contact us for more details, and you will receive fast and professional assistance.
References
- Wang, Xin, et al. "Mass spectrometry analysis of S-nitrosylation of proteins and its role in
cancer, cardiovascular and neurodegenerative diseases." TrAC Trends in Analytical Chemistry
(2022): 116625.
- Stomberski, Colin T., Douglas T. Hess, and Jonathan S. Stamler. "Protein S-nitrosylation: determinants of
specificity and enzymatic regulation of S-nitrosothiol-based signaling." Antioxidants & redox
signaling 30.10 (2019): 1331-1351.
- Zhao, Qian, et al. "Recent advances in predicting protein S-nitrosylation sites." BioMed
Research International 2021 (2021).
Our products and services are for research use only.

 Fig. 1 A schematic
diagram of protein S-nitrosylation sites. (Zhao, Qian, et al., 2021)
Fig. 1 A schematic
diagram of protein S-nitrosylation sites. (Zhao, Qian, et al., 2021)