Protein tyrosine sulfation is a common protein post-translational modification (PTM) that has been reported for a
long time. In recent years, the significant role of tyrosine sulfation in physiological conditions and diseases has
received increasing attention. Based on extensive experience in protein PTM analysis and advanced platforms,
Creative Proteomics offers reliable and customized tyrosine sulfation analysis services to
accelerate customers' projects in a comprehensive manner. Identification of new protein sulfation sites may be
essential and will contribute to a better understanding of the function of tyrosine sulfation.
Protein tyrosine sulfation
Since up to 1% of total protein tyrosine residues in an organism can be sulfated, sulfation is one of the most
abundant PTMs of tyrosine (Tyr). Tyr sulfation is also an enzyme-catalyzed modification that involves the transfer
of a sulfo group from a 3'-phosphoadenosine-5' phosphosulfate (PAPS) donor to the hydroxyl group of a Tyr
residue via a tryrosylprotein sulfotransferase. Sulfation of tyrosine residues occurs mainly in the Golgi apparatus;
therefore, secreted and extracellular proteins may be sulfated as they pass through the Golgi apparatus.
Tyrosine-sulfated proteins have been discovered in all animals (from lower invertebrates up to humans) and the plant
kingdom (from lower to higher plants). Although occurring throughout metazoan evolution, tyrosine-sulfate proteins
have been shown to be absent in unicellular eukaryotes and prokaryotes, suggesting that this PTM is associated with
some aspects of multicellular organisms. Protein tyrosine sulfation has been reported to be associated with a range
of biological processes such as cell signaling, molecular recognition, hormone regulation, viral entry into host
cells, and as a key factor in many diseases involving autoimmune responses, HIV infection, lung diseases, multiple
sclerosis, and cellular enzyme regulation.
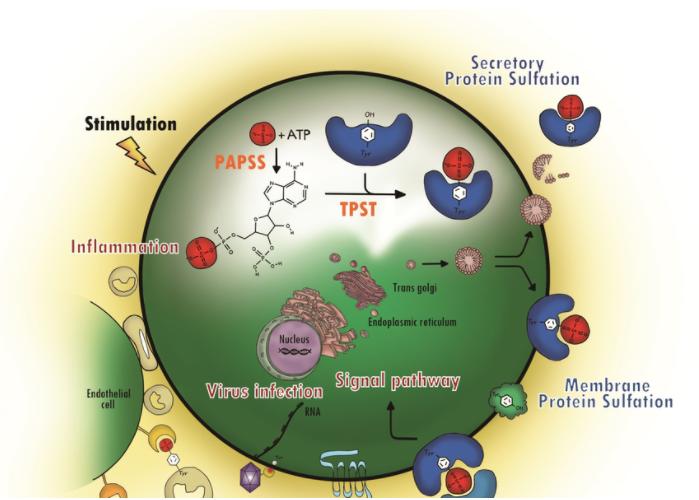 Fig. 1
Protein tyrosine sulfation (PTS) and its biological path. (Yang, Yuh-Shyong, et al., 2015)
Fig. 1
Protein tyrosine sulfation (PTS) and its biological path. (Yang, Yuh-Shyong, et al., 2015)
Proteomics analysis of sulfation
To meet the identification of tyrosine-sulfated proteins in various complex samples, we have utilized a series of
highly sensitive methods, including radioactive labeling, amino acid labeling, and mass spectrometry (MS). Among
these methods, the radioactive labeling method for tyrosine-sulfated protein identification consists of labeling
with radioactive sulfate followed by tyrosine sulfate analysis of a given protein. The sulfation of tyrosine
residues in proteins will be demonstrated using reversed-phase high-performance liquid chromatography (RP-HPLC) and
a pure protein or the use of specific tyrosine sulfate antibodies is usually required.
- MS-based tyrosine sulfation analysis
The MS-based analysis is a major method for studying the PTM of tyrosine residues. Since the sulfopeptides are
particularly difficult to analyze by the standard MS method, we focus on sample conditions, fragmentation methods,
and ionization parameters to improve the analysis workflow and ensure a reliable study of protein sulfation and a
full understanding of the protein sulfation state.
Our sample preparation consists of two main parts, namely protein extraction and digestion. We accept a variety of
samples such as protein extracts, cell samples, and tissue samples. It is necessary to note that please consult our
technical staff before sending samples. Importantly, we will test the samples you provide before the official
experiment.
- Tyrosine sulfation site prediction
We have strong expertise in protein Tyr sulfation analysis as well as tyrosine sulfation site prediction. Based on a
robust and novel platform for sulfation site identification, we are able to provide efficient services to meet the
specific needs of our customers worldwide. Our Tyr sulfation site prediction service combines protein secondary
structure, amino acid physicochemical properties, and residue sequence order information to predict Tyr sulfation
sites from the primary sequence of a protein.
Major benefits of our service
- Advanced equipment and platform.
- Optimized analysis workflow.
- Highly sensitive methods are available for the analysis of Tyr sulfation of proteins.
- Customized services from experienced experts.
If you are interested in our service, please contact us for more details.
References
- Yang, Yuh-Shyong, et al. "Tyrosine sulfation as a protein post-translational modification."
Molecules 20.2 (2015): 2138-2164.
- Rodríguez-Paredes, Manuel, and Frank Lyko. "The importance of non-histone protein methylation in cancer
therapy." Nature reviews Molecular cell biology 20.10 (2019): 569-570.
Our products and services are for research use only.

 Fig. 1
Protein tyrosine sulfation (PTS) and its biological path. (Yang, Yuh-Shyong, et al., 2015)
Fig. 1
Protein tyrosine sulfation (PTS) and its biological path. (Yang, Yuh-Shyong, et al., 2015)