Proteins, despite their small molecular stature, are indispensable elements within the cellular milieu. They play an integral role in myriad life-sustaining activities within cells, achieving this through a series of post-translational modifications. This modus operandi aids in regulating physiological processes and signal transduction within cells. Among these, protein phosphorylation, a unique mode of protein modification, endows proteins with novel characteristics and functions.
Via phosphorylation, the structural configuration of a protein can be altered, thereby influencing the intracellular signal transduction network. This consequential alteration embodies a pivotal function in various biological aspects such as gene transcription, expression, cell proliferation, differentiation, apoptosis, signal transduction, immunomodulation, tumorigenesis, and more. This intricate process is analogous to an elaborate dance; every single movement, each turn, is choreographed meticulously by phosphorylation.
Introduction to Protein Phosphorylation
Protein phosphorylation, a biochemical process, comes to life through enzymatic action catalyzing the transfer of phosphate groups from the γ position of ATP to the amino acid side chains of proteins. This process has the capacity for reversal, in which the enzyme that stimulates protein phosphorylation is referred to as Protein Kinase (PK), while the enzyme that propels the dephosphorylation of proteins is designated as Protein Phosphatase (PPase) (Figure 1). As a pivot mechanism, protein phosphorylation plays a paramount role in managing and controlling the activity and functionality of proteins.
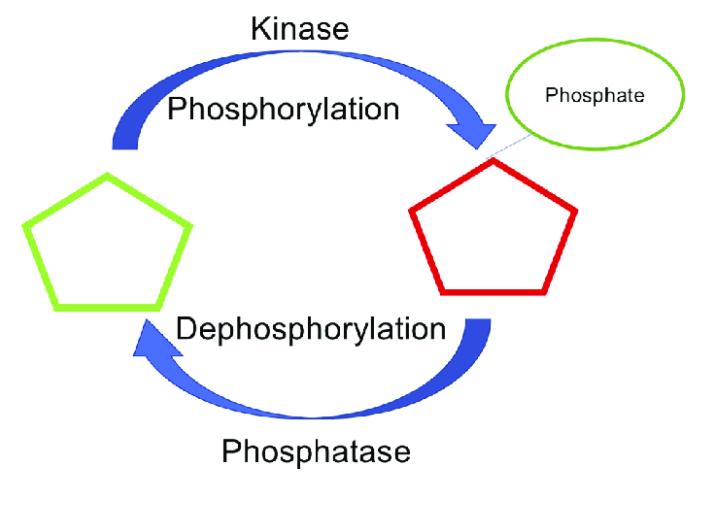 Figure 1 Phosphorylation and dephosphorylation (Meenal Chaudhari et al,.2021)
Figure 1 Phosphorylation and dephosphorylation (Meenal Chaudhari et al,.2021)
Protein Phosphorylation Type and Sites
Protein phosphorylation is a ubiquitous regulatory mechanism within biological organisms. According to research, over 90% of approximately 21,000 proteins encoded by the human genome undergo phosphorylation. Depending on the difference in phosphorylated amino acid residues, protein phosphorylation can be divided into four major categories. Detailed classification and phosphorylation sites are shown in Table 1, while the molecular formulae of the nine phosphorylated amino acids are depicted in Figure 2. Phosphorylation on serine, threonine and tyrosine residues accounts for more than a third of these categories: serine residues are the most common (86.4%), followed by threonine residues (11.8%), with tyrosine residues being relatively less frequent (1.8%). The phosphorylation of serine, threonine, and tyrosine constitutes O-phosphorylation. O-phosphorylation shows high stability under acidic conditions and therefore receives extensive attention in the fields of cell biology and phosphoproteomics. However, due to the unstable nature of N-phosphorylation, the detection is extremely challenging. Moreover, the scarcity of protein kinases and phosphatases catalysing N-phosphorylation further hinders research in this area. Comparative to the other two types of phosphorylation (S-phosphorylation and acyl-phosphorylation), there is less reported research.
Table1: Protein Phosphorylation Types and sites
| Protein Phosphorylation Types |
Protein Phosphorylation Sites |
Bond Between Amino Acid and Phosphate Group |
| O-Phosphorylation |
Serine (Ser; S); Threonine (Thr; T); Tyrosine (Tyr; Y) |
Phosphoester bond P-O |
| N-Phosphorylation |
Histidine (His; H); Arginine (Arg; R); Lysine (Lys; K) |
Amino acid phosphate ester bond P-N |
| S-Phosphorylation |
Aspartic acid (Asp; D); Glutamic acid (Glu; E) |
Phosphoanhydride bond P-O |
| Acyl Phosphorylation |
Cysteine (Cys; C) |
Thiophosphate ester bond P-S |
Protein Phosphorylation Related Enzymes
The human genome is enriched with approximately 500 kinds of kinases and more than 200 phosphatases, collaboratively mediating the process of protein phosphorylation, thereby manifesting this process as notably intricate. Within this assembly of phosphorylation-associated enzymes, they play pivotal roles in the nuanced regulation of numerous cellular processes within an organism.
Protein kinases, a member of the kinase family, serve as phosphotransferases. Their function resides in transferring the phosphate group from the gamma position of ATP to the specific amino acids of substrate proteins, hence actualizing protein phosphorylation and further underscored its physiological and biochemical functionalities. In the human genome, around 2% of human genes encode 518 protein kinases. Based on the divergences of phosphorylated amino acid residues, protein kinases can be categorised into five major classes: serine/threonine protein kinases, tyrosine protein kinases, histidine/lysine/arginine protein kinases, cysteine protein kinases, and aspartic acid/glutamic acid protein kinases.
In contrast to the function of protein kinases, protein phosphatases are responsible for the dephosphorylation of phosphorylated proteins. Currently, approximately 226 known protein phosphatases are being recognised. Determined by structural characteristics, protein phosphatases can be segregated into three families: protein phosphatase family, metal-dependent protein phosphatase family, and protein tyrosine phosphatase family.
Methods of Protein Phosphorylation Detection
In the domain of proteomics research, the analysis of protein phosphorylation alongside the identification of phosphorylation sites has emerged as a focus. The current strategies for detecting protein phosphorylation encompass techniques such as the use of specific antibodies (like western blot), radiolabeling with P32, mass spectrometry, integration of chemistry with biology, and nuclear magnetic resonance, among others. Each technique carries its individual strengths and limitations (refer to Table 2 for details), allowing researchers to choose the most suitable method in accordance with the requirements of their experimentation. More potential strategies for the detection of protein phosphorylation can be observed in the article "Methods of Protein Phosphorylation Detection .
Mass spectrometry phosphoproteomics
Utilizing mass spectrometry technology, we are able to assess the primary structure of proteins, including their molecular weight, sequences of amino acids in polypeptides, and the number and positions of polypeptides or disulfide bonds. This can facilitate the detection and identification of post-translational modifications. Mass spectrometry predominantly operates on the basis of two principles: Firstly, the relative molecular weight of a peptide segment modified with a single phosphate group increases by 79.983, compared to a non-phosphorylated peptide segment. Secondly, phosphorylated peptides generate a signature fragment ion useful for identification, a phenomenon not seen with non-phosphorylated modifications.
In theory, protein phosphorylation can be detected by mass spectrometry. However, when using mass spectrometry to analyze proteolytically digested phosphorylated proteins, it may not detect the entire protein sequence, potentially overlooking significant phosphorylated regions. Moreover, within cells, the amount of phosphorylated proteins is relatively low, and the signals generated by phosphorylated peptide segments can easily be suppressed by signals from non-phosphorylated peptide segments. Therefore, it is essential to enrich phosphorylated proteins and peptide segments prior to mass spectrometry analysis.
Several enrichment methods have been established for phosphorylated protein and peptide segments, including: (1) two-phase phosphorylated peptide maps; (2) high-resolution gel electrophoresis; (3) reverse-phase high-performance liquid chromatography; (4) metal oxide affinity chromatography or solid phase metal affinity chromatography; (5) high-affinity antibody immunoprecipitation.
Phosphoproteomics applications
The phosphorylation and dephosphorylation of proteins represent the most ubiquitous and crucial regulatory mechanisms that modulate the vigor and functionality of proteins. The study of these mechanisms is therefore of significant importance and applications have been extended to a broad range of research fields involving microbiology, botany, and zoology.
In the first instance, the study of these modifications can be instrumental within the investigation of cellular signal transduction networks. By analyzing alterations at the proteomic level in phosphorylated proteins, it's feasible to identify key factors within critical signaling pathways, thereby revealing the network structure and dynamic changes induced by phosphorylation control.
Additionally, phospho-proteomics may serve as a valuable tool for early disease diagnosis and treatment. The analysis of phospho-proteomic data can facilitate the identification of proteins related to specific diseases, offering a fresh perspective on new targets for early disease diagnosis, as well as pathways for drug development.
Lastly, phospho-proteomics can expedite drug screening and evaluation processes. Many therapeutic drugs achieve their desired effects by intervening in protein phosphorylation modifications. Through the analysis and evaluation of phospho-proteomics, it is viable to screen and appraise drug molecules associated with specific diseases, thus helping to accelerate the process of drug discovery and development.
Table 2: Comparison of protein phosphorylation detection methods
| Method |
Principle |
Merits |
Shortages |
| Mass spectrometry |
m/z based on fragments technologies |
Qualitative and quantitative analysis of phosphorylation;Screen inhibitors |
Fussy sample processing and time-consuming; |
| Specific antibodies |
Antibody immunoaffinity to phosphorylated amino acids |
Quantitative analysis level of phosphorylation |
Hard to generate site-specific phosphorylation antibodies, especially for N-phosphorylation; |
| Radionucleotide incorporation |
Autoradiograph of ³2p from phosphate donor [γ-³2p] ATP |
Qualitative and quantitative analysis of phosphorylation in combination with mutation experience; |
Time-consuming;standard laboratory conditions; |
| Chemical biology strategies |
Incorporation or chemical ligation of unnatural amino acid at interested site |
Exploit the role of unnatural amino acid in protein |
Limited to large proteins;required to mutate the desired site in a protein;probably hard to refold after protein denaturation; |
| NMR spectroscopy |
Changes in chemical shifts response to phosphorylation |
Qualitative and quantitative analysis of cluster phosphorylation and acid-labile PTMs; Monitoring the phosphorylation cascades in a noninvasive and nondestructive way;Screen inhibitors; |
Isotope labeled protein samples; |
(B. Huang et al. International Journal of Biological Macromolecules 145 .2020)
References
- Olsen JV, Blagoev B, Gnad F, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635-648.
- Tape CJ, Worboys JD, Sinclair J, Gourlay R, Vogt J, McMahon KM, Trost M, Lauffenburger DA, Lamont DJ, Jørgensen C. Reproducible automated phosphopeptide enrichment using magnetic TiO2 and Ti-IMAC. Anal Chem. 2014 Oct 21;86(20):10296-302. doi: 10.1021/ac5025842. Epub 2014 Oct 2. PMID: 25233145; PMCID: PMC4206527.
- Sharma K, D'Souza RCJ, Tyanova S, et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell reports. 2014;8(5):1583-1594.
- Palumbo AM, Smith SA, Kalcic CL, Dantus M, Stemmer PM, Reid GE. Tandem mass spectrometry strategies for phosphoproteome analysis. Mass Spectrom Rev. 2011 Jul-Aug;30(4):600-25. doi: 10.1002/mas.20310. Epub 2011 Feb 3. PMID: 21294150.
- Hennrich ML, Romanov N, Hornung J, et al. The proteome-wide spectrum of targets for C-kinase/PKC isoforms. eLife. 2018;7.
Our products and services are for research use only.


 Figure 1 Phosphorylation and dephosphorylation (Meenal Chaudhari et al,.2021)
Figure 1 Phosphorylation and dephosphorylation (Meenal Chaudhari et al,.2021)