The Importance of Post-translational Modifications
Post-translational modifications (PTMs) play an indispensable role in biological organisms. These modifications introduce further complexity into protein structures, enhance their functionality, allow for more precise regulation, and delegate specific roles to these proteins. PTMs commonly include ubiquitination, phosphorylation, glycosylation, lipidation, methylation, and acetylation, among others.
Ubiquitination, in particular, plays a crucial role in physiological processes such as cell differentiation and apoptosis, DNA repair, immune response, and stress responses. Phosphorylation is involved in numerous physiological and pathological processes, including cellular signal transduction, neural activity, muscle contraction, cell proliferation, development, and differentiation. Glycosylation plays an integral part in various biological processes such as immune defense, viral replication, cell growth, and the provocation of inflammation. Lipidation is instrumental for intracellular signal transduction processes. Furthermore, methylation and acetylation of histones are implicated in transcriptional regulation.
Within the biological system, these post-translational modification processes do not exist in isolation. Instead, they intertwine and interact in intricate ways, highlighting an exquisite coordination in cellular functioning. This therefore underscores the integral roles PTMs play in life's complexity and diversity.
In recognition of their revelation of the mechanism of ubiquitin-regulated protein degradation, Israeli scientists A. Ciechanover, A. Hershko, and American scientist O. Rose were awarded the 2004 Nobel Prize in Chemistry. Their groundbreaking research steer the direction of protein degradation studies, facilitating further understanding of our immune system, holding immense significance in DNA repair and control, and the treatment of human diseases.
Again, ubiquitination is indeed a type of post-translational protein modification, a process involving the covalent modification of individual amino acid residues on proteins after translation from mRNA. The completion of the Human Genome Project symbolizes one of the greatest scientific achievements of the 20th Century. Detailed study of the Human Genome reveals that there are only about 30,000-50,000 genes, just 3-5 times that of the chromosomal genes in nematodes and fruit flies. Such a small number of genes cannot fully meet the regulatory needs of complex life processes in an organism, thus underlining the importance of post-translational protein modifications. These modifications render the protein structure more complex, its functions more comprehensive, regulations more precise, and actions more specific. Many physiological functions within a cell, such as responses to external environments, are achieved through dynamic post-translational protein modifications. The complexity of human life processes is not solely a result of direct gene expression; the possibility of a sole gene corresponding to multiple proteins - facilitated by post-translational modifications - further adds to this complexity.
Interplay and Influence among Various Post-Translation Modifications
In the organism, post-translational modifications do not operate in separation. In many cellular activities, proteins modified through various post-translational processes collectively function. For instance, in signal transduction, external cell information receptors and corresponding responders (usually glycosylated proteins) on the cell membrane exterior interact with their respective ligand. These glycoproteins introduce stimuli signals from the cell's environment into the cell membrane, primarily transmitting it to associated lipoproteins. Simultaneously, in most signal transduction processes, lipoproteins typically initiate a series of protein phosphorylation, each of which is regulated by particular kinases and comprises the primary actor in the signal transduction process.
Multiple PTMs for a Single Protein
A single protein may undergo multiple post-translation modifications. Diverse PTMs interact and harmonize with each other. Phosphorylation and glycosylation share many characteristics, particularly in their kinetics and pervasiveness within cells, existing in transcription factors, oncogenic products, and enzymes. Current data suggest the addition and removal of N-acetyl glucosamine operates as a regulatory mechanism within cells. Not only do phosphorylation and glycosylation share substantial similarities, but they also act reciprocally. In the gene expression process governed by RNA polymerase II, phosphorylation and glycosylation have distinctive modification roles. The C-terminal domain of RNA polymerase is dramatically glycosylated, swiftly and fully deglycosylated, and simultaneously phosphorylated as the polymerase enters the nucleus, interacts with transcription factors. This implies that a large number of acetyl glucosamine groups might exist on RNA polymerase only when it's non-phosphorylated. Phosphorylation and glycosylation occur on the internal and external sides of cell membranes, respectively, suggesting that glycosylation could share phosphorylation site and affect the degree of phosphorylation.
The axonal microtubule-associated protein Tau also undergoes extensive glycosylation, having over twelve glycosylation sites and typically containing four GlcNAc modification sites per molecule. In Alzheimer's patients' brains, Tau protein forms a hyperphosphorylated tangle of paired helical filaments. The phosphorylation of Ser262 residue, which hampers Tau protein from binding with microtubules, should be glycosylated under normal conditions. Hence, it is hypothesized that abnormal phosphorylation at this position arises from the deficiency of glycosylation.
Post-translational Modifications of Histones
Histones can be co-modified by methylation and acetylation. The main sites of acetylation and methylation on histones are the conservative lysine residues at the ends of histones H3 and H4. Histone acetylation modifications span the entire cell cycle, while methylation modifications mostly occur during G2 phase and during the assembly of heterochromatin. Experimental evidence shows that the acetylation and methylation of lysine residues at the histone ends have a unique association, which may have antagonistic or cooperative biological functions. For instance, during the preparation of chromatin with transcriptional activity, highly acetylated H4 is the preferred substrate for the methylation of histone H3, suggesting that these modifications could work together to promote transcription. However, the operational mechanisms are still unclear.
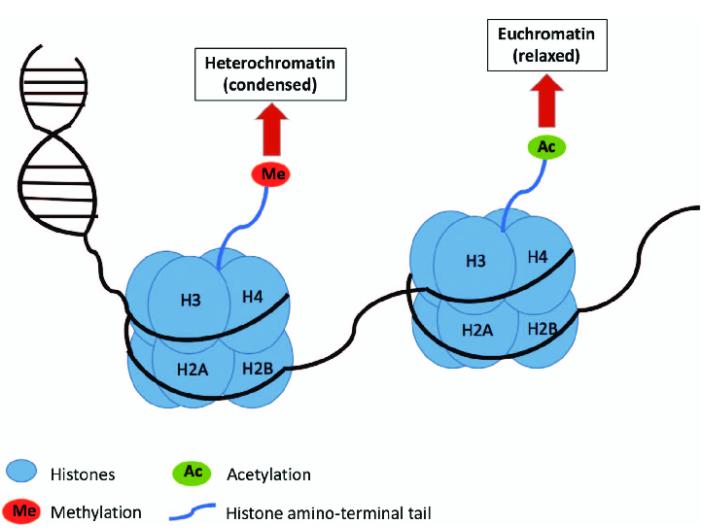 Histone methylation and acetylation (Somi Kim et al,. 2017)
Histone methylation and acetylation (Somi Kim et al,. 2017)
The Future of Protein Post-Translational Modification
Since protein post-translational modifications are not directly determined by genes, studying these modifications have profound implications for proteomics research, culminating in the birth of "post-translational modification proteomics." Post-translational modification proteomics is currently a hot research topic internationally. Investing in post-translational modification proteomics not only helps understand the critical significance of post-translational modifications in life processes but also provides substantial assurance for future drug development. Identifying aberrant molecular targets in abnormal cells will facilitate understanding how protein interactions are regulated by post-translational modification processes. Understanding the factors that regulate post-translational modification processes will contribute to revealing cellular processes and protein network functions at the molecular level, ultimately guiding more accurate molecular-targeted drug control.
References
- Venter J C, Adams M D, Myers E, et al. The sequence of the human genome. Science, 2001, 291: 1304~1351
- Rice J C, Allis C D. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Current Opinion in Cell Biology, 2001, 13: 263~273
- Benjamin G D. Mimicking posttranslational modifications of proteins. Science, 2004, 303: 480~482
Our products and services are for research use only.


 Histone methylation and acetylation (Somi Kim et al,. 2017)
Histone methylation and acetylation (Somi Kim et al,. 2017)