Protein post-translational modifications (PTMs) represent a crucial regulatory mechanism present in both animal and plant systems. PTMs encompass a diverse array of modifications primarily facilitated by specific enzymes that attach or remove functional groups on amino acid side chains, thereby modulating protein function. In plants, there exists a wide spectrum of modification types. According to the Plant PTM Viewer, 33 distinct PTMs have been identified in plants, involving 203,744 proteins and 585,169 modification sites (as depicted in Figure 1). PTMs can induce significant functional changes in proteins without altering their expression levels. These modifications can precisely regulate protein structure, function, and protein-protein interactions, thereby playing a vital role in various biological processes.
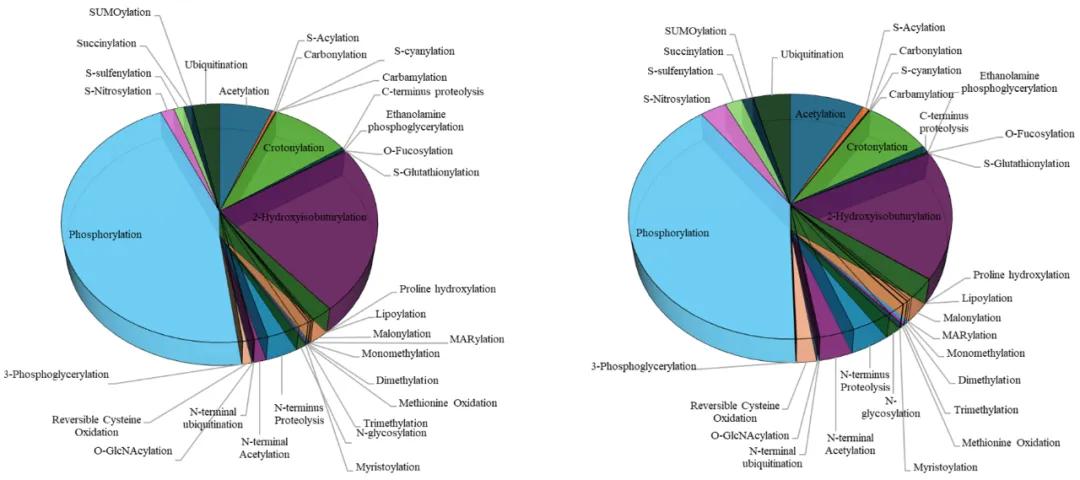 Figure 1 Plant PTM Viewer statistics of 33 PTMs in plants (Plant PTM Viewer website). (A) Modification sites of 33 PTMs in plant species; (B) Proteins of 33 PTMs in plants.
Figure 1 Plant PTM Viewer statistics of 33 PTMs in plants (Plant PTM Viewer website). (A) Modification sites of 33 PTMs in plant species; (B) Proteins of 33 PTMs in plants.
Varied Research Subjects in PTMs
During the process of PTMs, various modifying enzymes add or remove specific functional groups on their substrate proteins. Based on the nature of these functional groups, PTMs can be classified into several types, including phosphorylation, acetylation, crotonylation, succinylation, butyrylation, ubiquitination, glycosylation, S-nitrosylation, and propionylation. These modification processes necessitate the interaction between modifying enzymes and their respective substrates. Consequently, research on PTMs typically focuses on either the modifying enzymes or the substrates. In the subsequent sections of this paper, specific research methodologies will be elucidated, and the analysis of the modification relationships between enzymes and their substrates, following their identification, will be discussed.
Modifying Enzymes in PTMs
PTMs involve a diverse array of enzymes, including kinases, phosphatases, transferases, and ligases. These enzymes play pivotal roles in the modification processes, with some acting as primary modifiers and others facilitating the modification. In contemporary PTM research, significant emphasis is placed on modifying enzymes. Notably, certain modifications, such as phosphorylation, are reversible (as illustrated in Figure 2). Thus, reversible modifications require the involvement of both modifying enzymes (e.g., kinases) and demodifying enzymes (e.g., phosphatases). As demodifying enzymes also mediate the modification process, they are classified under the category of modifying enzymes.
Research on modifying enzymes can be conducted by screening interacting proteins and utilizing proteomic approaches specific to post-translational modifications. This paper will present relevant case studies based on these two approaches, further elucidating the critical roles of modifying enzymes in PTMs.
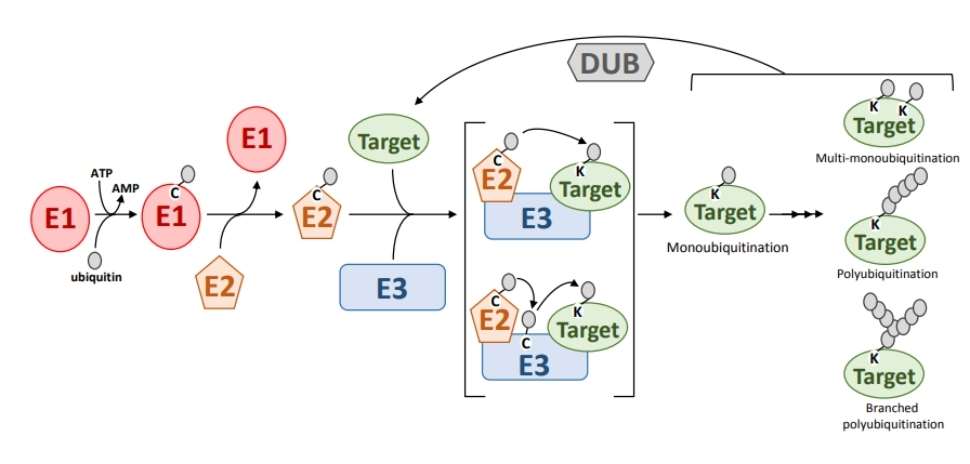 Figure 2 Reversible PTMs. The reversible process of ubiquitination.
Figure 2 Reversible PTMs. The reversible process of ubiquitination.
Methods for Screening Interacting Proteins
Various methods exist for the screening of interacting proteins, such as yeast two-hybrid screening and immunoprecipitation-mass spectrometry (IP-MS).
In March 2024, Hu Zhangjian's research group from Zhejiang University published a study in Plant Physiology titled "Phosphorylation of Sugar Transporter TST2 by Protein Kinase CPK27 Enhances Drought Tolerance in Tomato." The study aimed to elucidate how calcium-dependent protein kinase CPK27 enhances drought tolerance in tomatoes through the regulation of soluble sugar accumulation by identifying the target substrates of CPK27.
Interestingly, the authors identified the vacuolar membrane sugar transporter TST2 as an interacting protein of CPK27 using yeast two-hybrid screening (Figure 3A). Additionally, phosphoproteomic analysis revealed five phosphorylation sites on TST2 (Figure 3B). The concordance of results from both experimental approaches strongly suggests that TST2 is a likely phosphorylation substrate of CPK27.
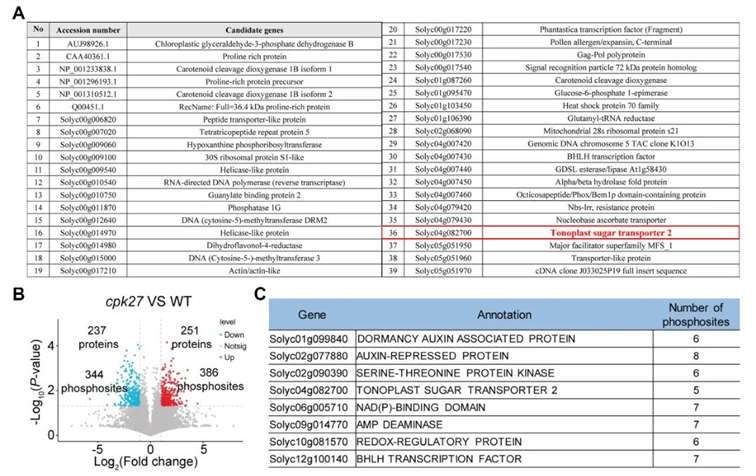 Figure 3 Screening and identification of CPK27 substrates (Zhu et al., 2024). (A) Screening of interacting proteins of CPK27 through yeast screening library; (B) Proteomic analysis of phosphorylation modifications of WT and cpk27 before drought stress; (C) More than 5 modifications identified in the phosphorylation modification proteomics Candidate substrates for the site.
Figure 3 Screening and identification of CPK27 substrates (Zhu et al., 2024). (A) Screening of interacting proteins of CPK27 through yeast screening library; (B) Proteomic analysis of phosphorylation modifications of WT and cpk27 before drought stress; (C) More than 5 modifications identified in the phosphorylation modification proteomics Candidate substrates for the site.
In this study, the authors employed two distinct methods to identify substrates of CPK27, both of which identified the same substrate. Some studies utilize only a single method for substrate screening; therefore, researchers may choose to use one method or both, depending on the specific requirements of their investigation.
Furthermore, it is posited that when the target protein is a modifying enzyme, employing proteomic approaches to identify PTMs is likely the optimal strategy. Proteomic analyses can reveal proteins with upregulated or downregulated modifications across different materials or treatments, along with the specific modification sites. This method provides more comprehensive information about potential substrates compared to yeast two-hybrid screening, thereby facilitating subsequent experimental steps.
Substrates of Modifying Enzymes
As previously noted, numerous proteins undergo modifications within plant cells. Given the crucial regulatory roles of these modifications in protein function, many studies investigate their impact on protein functionality. Notably, substrates of modifying enzymes encompass not only non-modifying enzyme proteins but also various modifying enzyme proteins. Furthermore, a single protein may undergo multiple types of modifications. In summary, different types of intracellular modifications collaborate to facilitate various complex physiological processes. Relevant discussions on this topic can be found in literature addressing Crosstalk Between Protein Post-Translational Modifications.
In plant gene function studies, when investigating modifications on a specific substrate protein and identifying its modifying enzymes, researchers can employ the method outlined in "Methods for Screening Interacting Proteins," as described earlier.
In February 2024, The Plant Cell journal published a research article titled "MAC3A and MAC3B mediate degradation of the transcription factor ERF13 and thus promote lateral root emergence." In previous investigations, the authors observed that high concentrations of auxin led to the degradation of the ethylene response factor ERF13. To delve deeper into this process, the authors aimed to identify the ubiquitin ligase interacting with ERF13, hypothesizing that ubiquitination might elucidate ERF13's degradation. Consistently, IP-MS successfully identified the candidate ubiquitin ligase MAC3B among ERF13's interacting proteins (Figure 4). Subsequent molecular experiments validated MAC3B's role in mediating the ubiquitination and subsequent degradation of ERF13.
 Figure 4 Screening of ERF13 interacting proteins by IP-MS (Yu et al., 2024).
Figure 4 Screening of ERF13 interacting proteins by IP-MS (Yu et al., 2024).
In many studies, when investigating the modifications on a particular substrate protein, not all research endeavors involve screening for modifying enzymes. Some studies solely identify the modification sites and subsequently establish the connection between the modification and the research focus through subsequent validation, as will be elaborated upon in the following sections.
However, for a comprehensive understanding of the molecular mechanisms underlying modifications, it is deemed optimal to ascertain both the modifying enzymes and substrates. The preceding two sections of this paper discussed methods for determining one entity through the other, whether modifying enzymes or substrates. Subsequently, the paper will delve into how to further investigate upon their determination to elucidate the relationship between the two modifications. Continued reading will provide insights into this matter.
Modifying Enzymes/Substrates
Protein-Protein Interactions
Upon identifying the modifying enzyme or substrate, further elucidating their interaction necessitates validation through various protein-protein interaction assays. Typically, confirming the upstream-downstream relationship between two conventional proteins requires intricate genetic experiments post interaction verification. However, for modifying enzymes and their substrates, the direction of modification is unidirectional, obviating the need for complex genetic experimentation to validate their relationship. Nonetheless, relevant experiments are still required to substantiate their modifying relationship.
In the previously mentioned research article "Phosphorylation of sugar transporter TST2 by protein kinase CPK27 enhances drought tolerance in tomato," the authors validated the interaction between CPK27 and TST2. Through yeast two-hybrid assays, pull-down assays, bimolecular fluorescence complementation (BiFC), and co-immunoprecipitation (Co-IP), they confirmed the interaction between CPK27 and TST2 (Figure 5), suggesting that CPK27 likely mediates the phosphorylation modification of TST2.
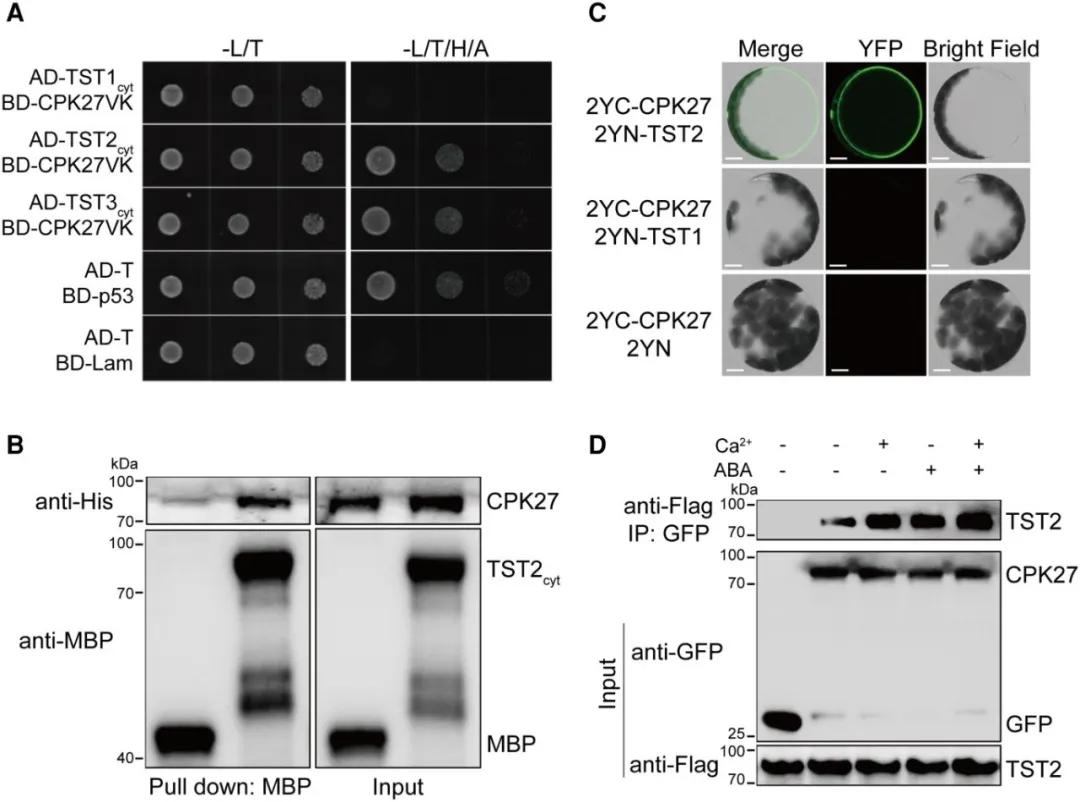 Figure 5 Verification of the interaction between CPK27 and TST2 through yeast two-hybrid, Pull-down, BiFC and Co-IP (Zhu et al., 2024).
Figure 5 Verification of the interaction between CPK27 and TST2 through yeast two-hybrid, Pull-down, BiFC and Co-IP (Zhu et al., 2024).
Validation of Modifications
In the process of detecting protein modifications, Western blot (WB) analysis using antibodies is the most common and effective method. However, for specific types of modifications, such as phosphorylation, due to their unique biochemical properties, besides utilizing specific antibodies to identify and detect modified proteins, one can also employ Phos-tag gels for WB separation and visualization of phosphorylated proteins, thereby enabling the detection of phosphorylation modifications. However, considering this article's main objective is to summarize and introduce general strategies and methods for protein modification, specific details regarding the use of Phos-tag gels for detecting phosphorylated proteins will not be extensively discussed. Interested readers are encouraged to consult relevant literature for further understanding.
Modified antibodies are primarily divided into two categories: pan-modified antibodies and site-specific modification antibodies. Pan-modified antibodies are mainly used to detect the modification state of a specific modification or a class of amino acid residues. However, they cannot precisely identify and detect the modification status of a specific amino acid residue in the protein. In contrast, site-specific modification antibodies have the ability to accurately detect the modification status of a specific amino acid residue in the protein. These two types of antibodies have different application scenarios and advantages in modification detection, meeting various research needs.
In the aforementioned research article "Phosphorylation of sugar transporter TST2 by protein kinase CPK27 enhances drought tolerance in tomato," after validating the interaction between CPK27 and TST2, the authors demonstrated the phosphorylation modification of TST2 using a phosphorylation pan-specific antibody (anti-phosphoserine/threonine) WB assay. Simultaneously, they detected a decreased level of phosphorylation modification of TST2 in the cpk27 mutant (Figure 6). Furthermore, after drought treatment, the difference in phosphorylation of TST2 between WT and cpk27 mutant was more pronounced (Figure 6), suggesting that TST2 may respond to drought conditions through CPK27-mediated phosphorylation modification.
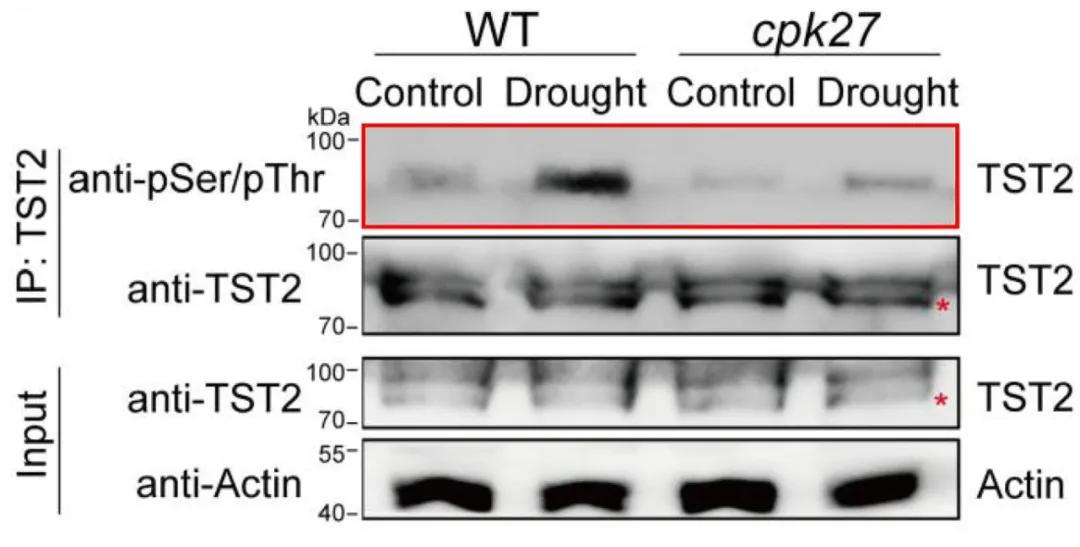 Figure 6 Detection of phosphorylation modifications on TST2 by phosphopan-antibody (anti-phosphorylated serine/threonine) (Zhu et al., 2024).
Figure 6 Detection of phosphorylation modifications on TST2 by phosphopan-antibody (anti-phosphorylated serine/threonine) (Zhu et al., 2024).
In November 2020, the journal Genomics, Proteomics & Bioinformatics published a research paper titled "Ubiquitinome Profiling Reveals the Landscape of Ubiquitination Regulation in Rice Young Panicles." The authors aimed to validate the reliability of protein ubiquitination modification proteomics. They employed ubiquitination pan-specific antibodies to detect the ubiquitination status of OsDRUS1 before and after mutation. The results indicated that in the mutant, there were scarcely observable bands representing OsDRUS1 modifications. However, evident modification bands were observed in materials stably expressing OsDRUS1-Myc (Figure 7). This observation confirms the occurrence of ubiquitination modification on OsDRUS1 and further corroborates the reliability of the proteomic findings.
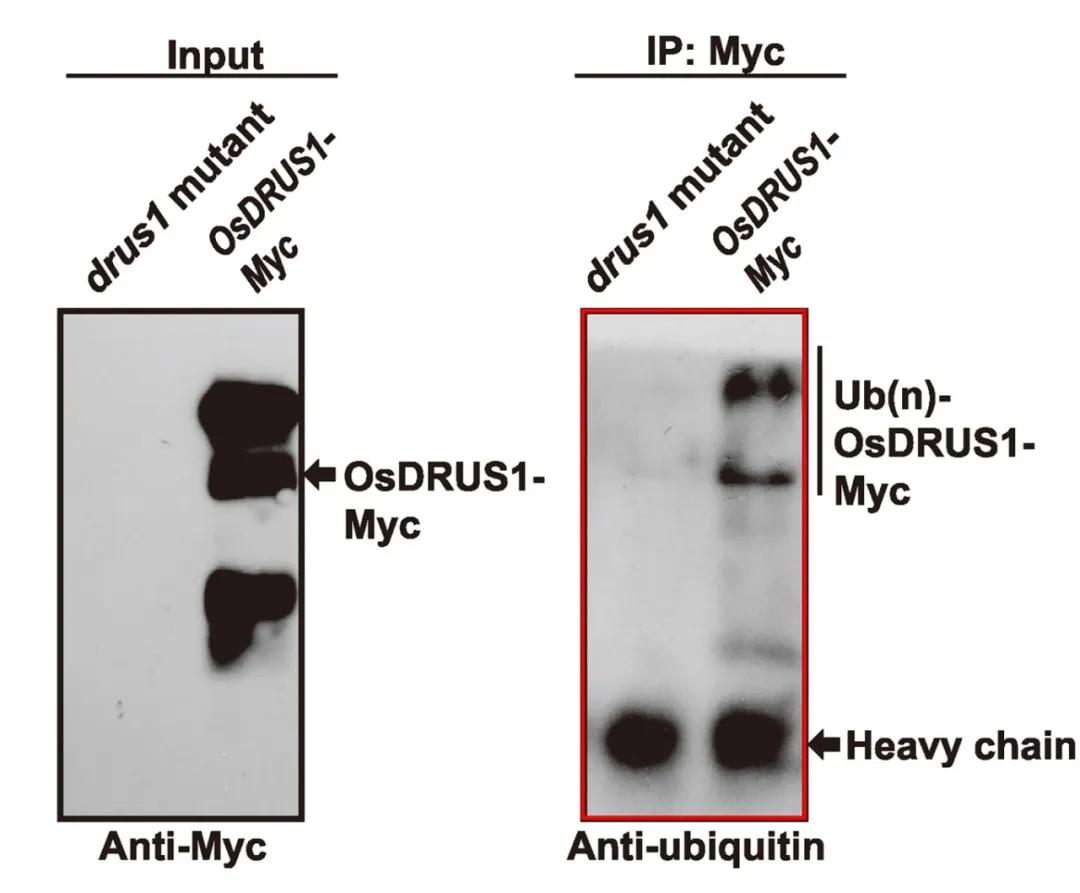 Figure 7 Detection of ubiquitination modification on OsDRUS1 through ubiquitination pan-antibody (Zhu et al., 2020).
Figure 7 Detection of ubiquitination modification on OsDRUS1 through ubiquitination pan-antibody (Zhu et al., 2020).
In the preceding sections, the detailed elucidation of protein modification detection using pan-specific antibodies has been provided. To deepen comprehension, this paper shares a series of practical cases where protein modifications were detected using site-specific antibodies. These cases aim to demonstrate the specific application and advantages of site-specific antibodies in protein modification detection, providing readers with richer and more practical references.
In October 2022, the journal Developmental Cell published a research paper titled "Root twisting drives halotropism via stress-induced microtubule reorientation." The authors aimed to validate whether phosphorylation on Serine 406 (Ser406) of SP2L is associated with salt stress. They performed Western blotting using site-specific phosphorylation antibodies targeting this residue and found that salt stress enhanced phosphorylation on the Ser406 residue. Moreover, this trend could be abolished by phosphatase treatment (Figure 8), indicating that phosphorylation on SP2L Ser406 residue is indeed regulated by salt stress.
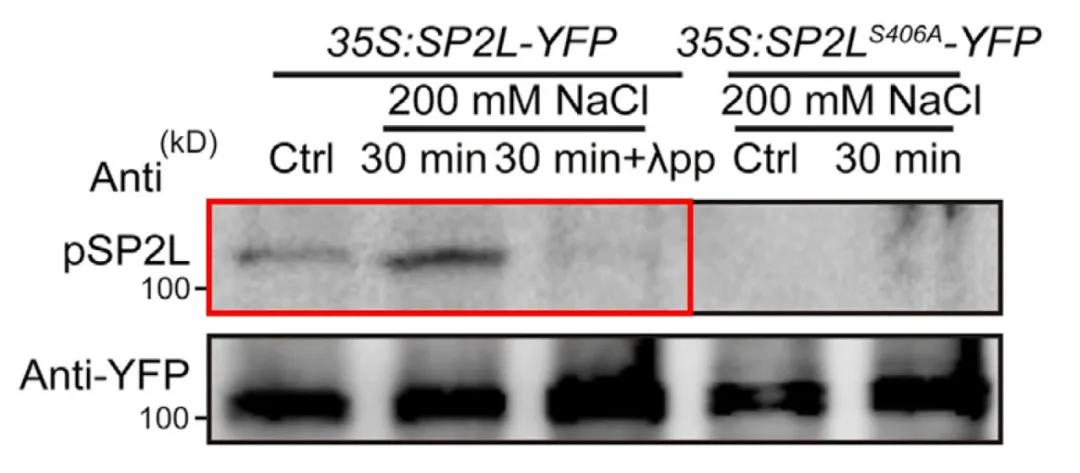 Figure 8 Detection of phosphorylation on SP2L by SP2L Ser406 phosphorylation antibody (Yu et al., 2022).
Figure 8 Detection of phosphorylation on SP2L by SP2L Ser406 phosphorylation antibody (Yu et al., 2022).
In September 2020, the journal Molecular Plant published a research paper titled "ESCRT-I Component VPS23A Is Targeted by E3 Ubiquitin Ligase XBAT35 for Proteasome-Mediated Degradation in Modulating ABA Signaling." To verify whether VPS23A undergoes ubiquitination-mediated degradation and to identify the ubiquitination sites, the authors conducted Western blotting using antibodies specific to lysine 48 (K48) and lysine 63 (K63) ubiquitination. Their findings revealed that VPS23A is primarily degraded through ubiquitination at the K48 residue (Figure 9).
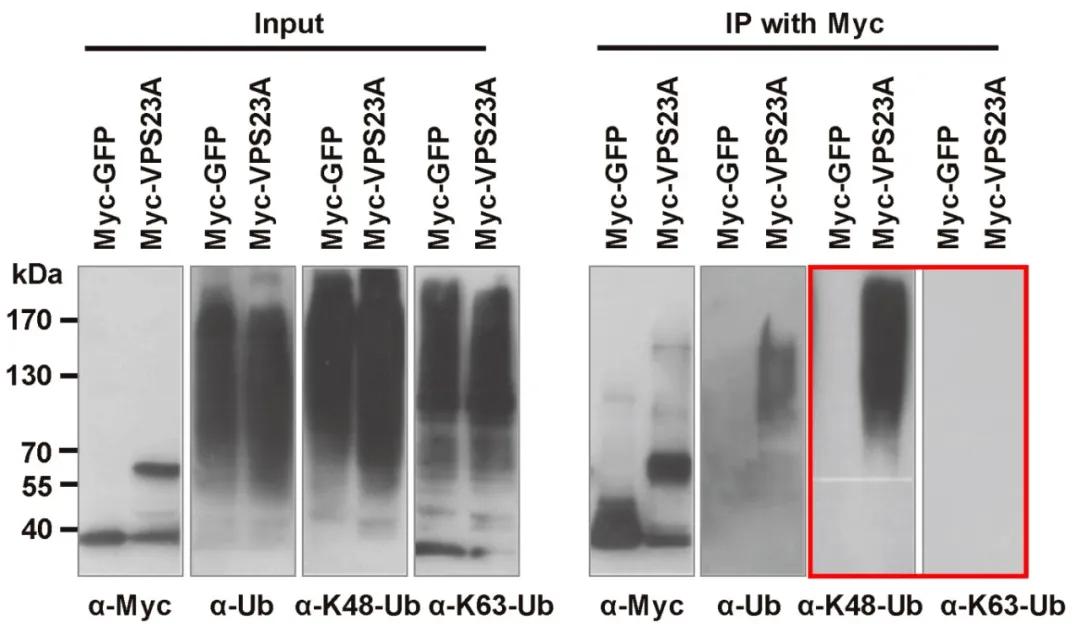 Figure 9 Detection of ubiquitination of VPS23A through K48 and K63 ubiquitination antibodies (Yu et al., 2020).
Figure 9 Detection of ubiquitination of VPS23A through K48 and K63 ubiquitination antibodies (Yu et al., 2020).
The preceding discussion pertains to technical approaches for detecting modification states within organisms. However, for extensively studied modifications like phosphorylation, analytical methods can also involve ex vivo assessments.
In March 2019, the journal Nature Plants published a research paper titled "Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase." To further substantiate the phosphorylation of PUX5, S40-7, ATG13, and eIF2B-δ1 by TOR (Target of Rapamycin) kinase, the authors conducted in vitro phosphorylation assays, confirming the phosphorylation of these substrates by TOR. Similarly, ubiquitination can also be simulated ex vivo, as demonstrated in previous discussions on "A Comprehensive Review of Protein Ubiquitination: Pathways, Site Predictions, Detection Methods, and Functions."
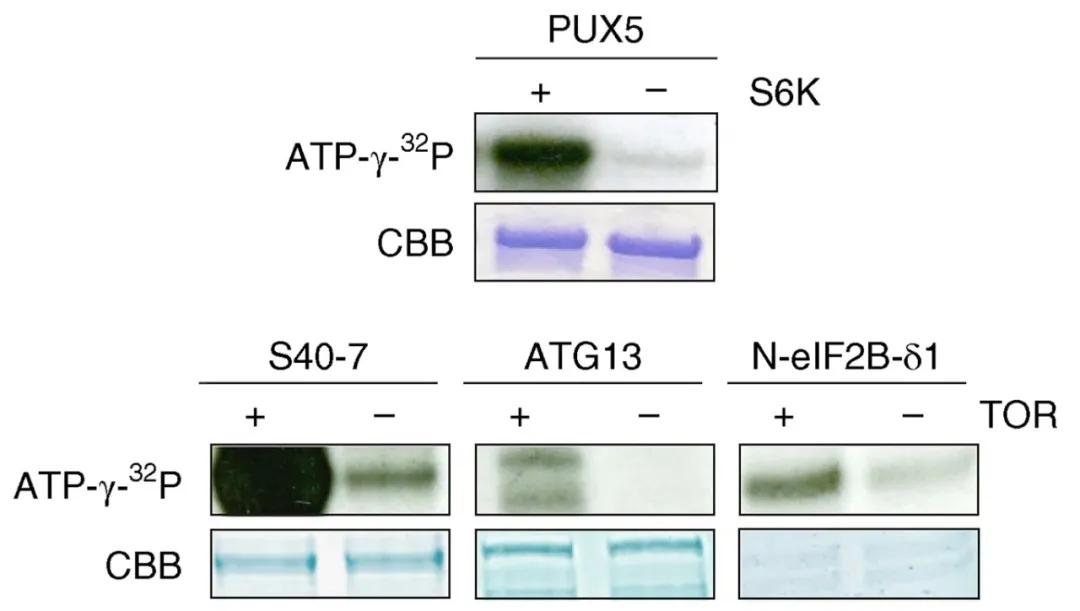 Figure 10 depicts an in vitro phosphorylation experiment (Van Leene et al., 2019). Substrates (PUX5, S40-7, ATG13, and eIF2B-δ1) were incubated with the kinase TOR and the phosphorylation donor (γ-32P ATP) under buffered conditions. Following termination of the reaction, SDS-PAGE electrophoresis was performed, and radioactive signals were detected using X-ray film exposure or a phosphorimaging system.
Figure 10 depicts an in vitro phosphorylation experiment (Van Leene et al., 2019). Substrates (PUX5, S40-7, ATG13, and eIF2B-δ1) were incubated with the kinase TOR and the phosphorylation donor (γ-32P ATP) under buffered conditions. Following termination of the reaction, SDS-PAGE electrophoresis was performed, and radioactive signals were detected using X-ray film exposure or a phosphorimaging system.
Identification of Modification Sites
Following the confirmation of the enzyme's capability to effectively modify the substrate, further efforts are often required to identify specific modification sites. Currently, two main methods are employed for identifying modification sites on substrates. Firstly, various prediction websites are utilized, leveraging existing research data to preliminarily infer modification sites. Secondly, immunoprecipitation (IP) techniques are employed to enrich substrates, followed by in-depth analysis using MS. It is noteworthy that prediction websites primarily focus on extensively studied modification types. This article compiles some commonly used prediction website resources for modification sites. However, given that this article has not comprehensively tested the specific prediction accuracy of each website, researchers should conduct practical experiments and validations based on their own requirements.
Table 1. Partial Protein Modification Site Prediction Websites.
Concerning prediction websites, it is acknowledged that they have inherent limitations regarding the types of modifications they cover, hence their predictive outcomes may not be entirely precise. To ensure result accuracy, the most reliable approach is to perform immunoprecipitation (IP) enrichment of substrates followed by MS analysis. This method resembles the IP-MS technique commonly used in identifying interacting proteins but with operational distinctions. In this approach, substrate proteins are solely enriched via IP, followed by in-depth analysis of the modification types and their corresponding sites on the substrate protein.
In September 2023, the journal Cell published a research paper titled "Amyloplast sedimentation repolarizes LAZYs to achieve gravity sensing in plants". To investigate how the key gravity-responsive factor LAZY mediates plant gravitropism, the authors conducted MS analysis of LAZY phosphorylation differences before and after gravity stimulation. They identified a total of 13 phosphorylation sites (Figure 11), with the phosphorylation levels of these sites generally upregulated after gravity stimulation, suggesting a potential association between plant gravitropism and LAZY phosphorylation.
 Figure 11 Identification of phosphorylation modification sites on LAZY by MS (Chen et al., 2023).
Figure 11 Identification of phosphorylation modification sites on LAZY by MS (Chen et al., 2023).
In November 2023, the journal Plant Physiology published a research paper titled "Protist ubiquitin ligase effector PbE3-2 targets cysteine protease RD21A to impede plant immunity." In order to identify the specific ubiquitination sites of the ubiquitin ligase PbE3-2 on RD21A, the authors performed mass spectrometry (MS) analysis after immunoprecipitation (IP) of RD21A. A total of 13 ubiquitination sites were identified through this analysis (Figure 12).
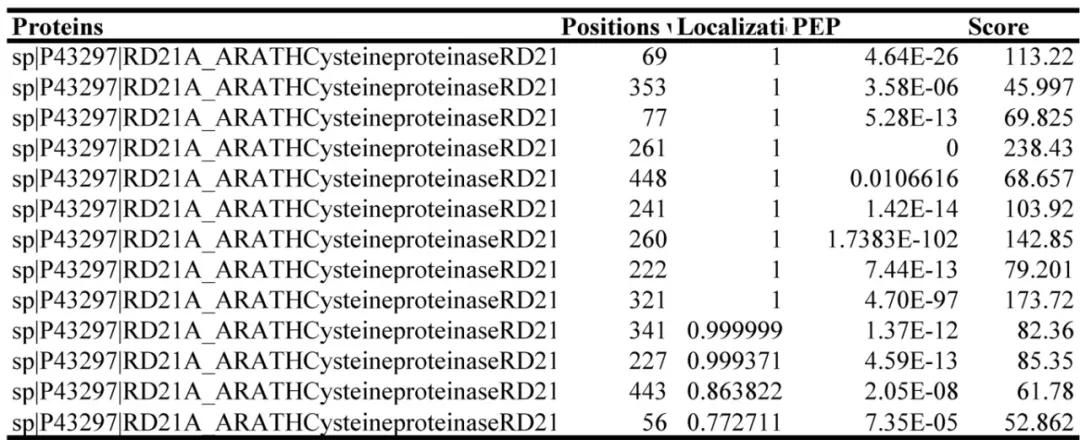 Figure 12 Identification of ubiquitination modification sites on RD21A by MS (Li et al., 2023).
Figure 12 Identification of ubiquitination modification sites on RD21A by MS (Li et al., 2023).
Site-Specific Verification
Following rigorous identification of specific modification sites, to further ascertain the occurrence of a modification at a given site, the method of site-specific verification can be employed. This involves mutating the amino acid at the modification site and verifying the modification status. Typically, if a particular amino acid residue of the substrate undergoes modification, mutating that residue to another type will decrease the level of modification on the substrate.
In the aforementioned study titled "Root twisting drives halotropism via stress-induced microtubule reorientation," upon confirming the phosphorylation modification on Ser406 residue of SP2L, researchers conducted site-specific verification experiments. Mutating Ser (serine) to Ala (alanine) resulted in the nearly complete disappearance of phosphorylation modification on SP2L, as shown in Figure 13. This outcome further confirms that phosphorylation of SP2L primarily occurs at the Ser406 residue, providing robust experimental evidence for related research endeavors.
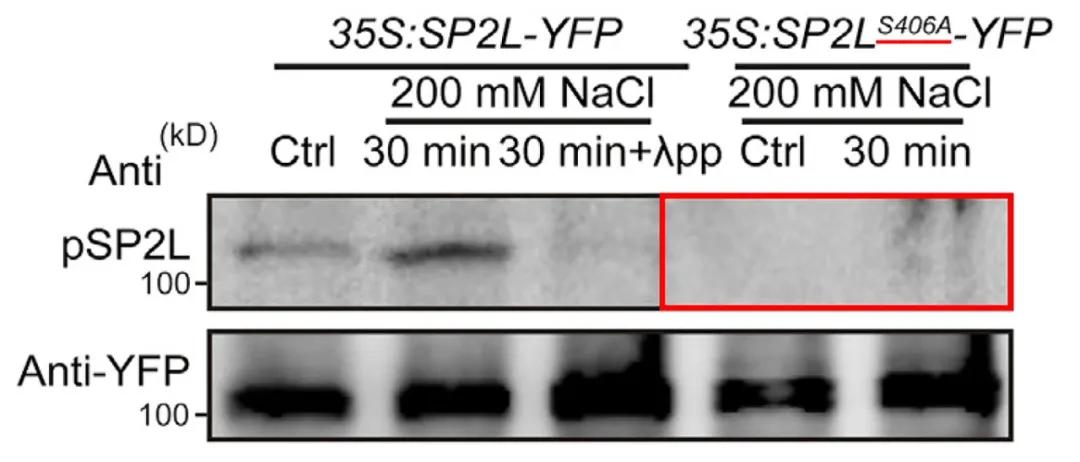 Figure 13 Phosphorylation status after mutation of SP2L phosphorylation site (Yu et al., 2022).
Figure 13 Phosphorylation status after mutation of SP2L phosphorylation site (Yu et al., 2022).
In this section, the paper will elaborate on how to further elucidate the relationship between modifying enzymes and substrates once they have been clearly identified. For certain novel modifications, given the current absence of corresponding antibodies for use, additional efforts and resources may be required to label modification groups effectively for detection. In essence, the core objective of various detection methods is to reveal differences in modification status among different materials, such as determining modification sites and evaluating modification levels. Ultimately, through in-depth analysis of the detection results, it will be possible to elucidate the significant role of these modifications in biological processes.
Summary
This paper aims to introduce research methods related to PTMs to the readers. It is important to emphasize that these methods mainly apply to the study scope of non-histone protein modifications, whereas discussions on histone protein modifications require other experimental techniques such as ChIP-seq and CUT&Tag. In recent years, PTMs have garnered significant attention, with their importance and research interest continually rising. Currently, research on PTMs primarily focuses on the exploration of individual modifications. However, as research progresses, the direction is gradually shifting from the study of single modifications to comprehensive investigations of multiple modifications, aimed at revealing the combined regulatory effects of different PTMs on proteins. Related articles such as "Crosstalk between post-translational modifications of proteins" delve into this in-depth. As one of the key factors contributing to protein diversity, the diverse types and functions of PTMs endow them with significant research value. It is imperative for researchers to delve deeper into unraveling the mysteries of PTMs, aiming to achieve more fruitful research outcomes in the field of life sciences.
References
- Li C, Luo S, Feng L et al. Protist ubiquitin ligase effector PbE3-2 targets cysteine protease RD21A to impede plant immunity. Plant Physiology, 2023, 194: 1764-1778.Chen J, Yu R, Li N et al. Amyloplast sedimentation repolarizes LAZYs to achieve gravity sensing in plants. Cell, 2023, 186: 4788-4802.e4715.
- McNulty DE, Sikorski TW, Annan RS In Analysis of Protein Post‐Translational Modifications by Mass Spectrometry 2016, p 17-87.
- Stone SL. Role of the Ubiquitin Proteasome System in Plant Response to Abiotic Stress. Int Rev Cell Mol Biol, 2019, 343: 65-110.
- Van Leene J, Han C, Gadeyne A et al. Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat Plants, 2019, 5: 316-327.
- Yu B, Zheng W, Xing L et al. Root twisting drives halotropism via stress-induced microtubule reorientation. Dev Cell, 2022, 57: 2412-2425.e2416.
- Yu F, Cao X, Liu G et al. ESCRT-I Component VPS23A Is Targeted by E3 Ubiquitin Ligase XBAT35 for Proteasome-Mediated Degradation in Modulating ABA Signaling. Mol Plant, 2020, 13: 1556-1569.
- Yu Z, Qu X, Lv B et al. MAC3A and MAC3B mediate degradation of the transcription factor ERF13 and thus promote lateral root emergence. The Plant Cell, 2024.
- Zhu C, Jing B, Lin T et al. Phosphorylation of sugar transporter TST2 by protein kinase CPK27 enhances drought tolerance in tomato. Plant Physiology, 2024.
- Zhu L, Cheng H, Peng G et al. Ubiquitinome Profiling Reveals the Landscape of Ubiquitination Regulation in Rice Young Panicles. Genomics Proteomics Bioinformatics, 2020, 18: 305-320.
Our products and services are for research use only.


 Figure 1 Plant PTM Viewer statistics of 33 PTMs in plants (Plant PTM Viewer website). (A) Modification sites of 33 PTMs in plant species; (B) Proteins of 33 PTMs in plants.
Figure 1 Plant PTM Viewer statistics of 33 PTMs in plants (Plant PTM Viewer website). (A) Modification sites of 33 PTMs in plant species; (B) Proteins of 33 PTMs in plants. Figure 2 Reversible PTMs. The reversible process of ubiquitination.
Figure 2 Reversible PTMs. The reversible process of ubiquitination. Figure 3 Screening and identification of CPK27 substrates (Zhu et al., 2024). (A) Screening of interacting proteins of CPK27 through yeast screening library; (B) Proteomic analysis of phosphorylation modifications of WT and cpk27 before drought stress; (C) More than 5 modifications identified in the phosphorylation modification proteomics Candidate substrates for the site.
Figure 3 Screening and identification of CPK27 substrates (Zhu et al., 2024). (A) Screening of interacting proteins of CPK27 through yeast screening library; (B) Proteomic analysis of phosphorylation modifications of WT and cpk27 before drought stress; (C) More than 5 modifications identified in the phosphorylation modification proteomics Candidate substrates for the site. Figure 4 Screening of ERF13 interacting proteins by IP-MS (Yu et al., 2024).
Figure 4 Screening of ERF13 interacting proteins by IP-MS (Yu et al., 2024). Figure 5 Verification of the interaction between CPK27 and TST2 through yeast two-hybrid, Pull-down, BiFC and Co-IP (Zhu et al., 2024).
Figure 5 Verification of the interaction between CPK27 and TST2 through yeast two-hybrid, Pull-down, BiFC and Co-IP (Zhu et al., 2024). Figure 6 Detection of phosphorylation modifications on TST2 by phosphopan-antibody (anti-phosphorylated serine/threonine) (Zhu et al., 2024).
Figure 6 Detection of phosphorylation modifications on TST2 by phosphopan-antibody (anti-phosphorylated serine/threonine) (Zhu et al., 2024). Figure 7 Detection of ubiquitination modification on OsDRUS1 through ubiquitination pan-antibody (Zhu et al., 2020).
Figure 7 Detection of ubiquitination modification on OsDRUS1 through ubiquitination pan-antibody (Zhu et al., 2020). Figure 8 Detection of phosphorylation on SP2L by SP2L Ser406 phosphorylation antibody (Yu et al., 2022).
Figure 8 Detection of phosphorylation on SP2L by SP2L Ser406 phosphorylation antibody (Yu et al., 2022). Figure 9 Detection of ubiquitination of VPS23A through K48 and K63 ubiquitination antibodies (Yu et al., 2020).
Figure 9 Detection of ubiquitination of VPS23A through K48 and K63 ubiquitination antibodies (Yu et al., 2020). Figure 10 depicts an in vitro phosphorylation experiment (Van Leene et al., 2019). Substrates (PUX5, S40-7, ATG13, and eIF2B-δ1) were incubated with the kinase TOR and the phosphorylation donor (γ-32P ATP) under buffered conditions. Following termination of the reaction, SDS-PAGE electrophoresis was performed, and radioactive signals were detected using X-ray film exposure or a phosphorimaging system.
Figure 10 depicts an in vitro phosphorylation experiment (Van Leene et al., 2019). Substrates (PUX5, S40-7, ATG13, and eIF2B-δ1) were incubated with the kinase TOR and the phosphorylation donor (γ-32P ATP) under buffered conditions. Following termination of the reaction, SDS-PAGE electrophoresis was performed, and radioactive signals were detected using X-ray film exposure or a phosphorimaging system. Figure 11 Identification of phosphorylation modification sites on LAZY by MS (Chen et al., 2023).
Figure 11 Identification of phosphorylation modification sites on LAZY by MS (Chen et al., 2023). Figure 12 Identification of ubiquitination modification sites on RD21A by MS (Li et al., 2023).
Figure 12 Identification of ubiquitination modification sites on RD21A by MS (Li et al., 2023). Figure 13 Phosphorylation status after mutation of SP2L phosphorylation site (Yu et al., 2022).
Figure 13 Phosphorylation status after mutation of SP2L phosphorylation site (Yu et al., 2022).