Current mechanistic research endeavors are dedicated to exploring various molecular dimensions, including genomics, epigenomics, transcriptomics, proteomics, and metabolomics, aiming to provide a comprehensive elucidation of biological processes through diverse technical approaches. Among these processes, post-translational modifications (PTMs) of proteins, such as phosphorylation, glycosylation, methylation, ubiquitination, lactylation, and palmitoylation, are pivotal. These modifications play central roles in regulating a myriad of biological processes within organisms. Notably, protein palmitoylation has emerged as a focal area of contemporary scientific investigation.
Protein Palmitoylation: An Overview
Palmitoylation is a significant post-translational modification involving the covalent attachment of a 16-carbon palmitoyl group to cysteine residues in proteins via a thioester bond. This modification markedly enhances the hydrophobicity of proteins, thereby impacting their subcellular localization, enzymatic activity, and trafficking within the endomembrane system. These effects contribute to the regulation of numerous biological processes.
Mechanistic Insights
The palmitoylation process is reversible. The addition of palmitoyl groups, known as palmitoylation, is mediated by members of the ZDHHC family of enzymes. In mammals, there are 23 ZDHHC members, numbered from ZDHHC1 to ZDHHC24, excluding ZDHHC10. Conversely, the removal of palmitoyl groups relies on a range of specific enzymes, including, but not limited to, APT1, APT2, PPT1, PPT2, ABHD17A/B/C, and ABHD10 (Figure 1).
Implications in Tumor Research
In the context of tumor research, palmitoylation serves as a critical modulator of protein function and localization, influencing various aspects of cancer biology. For instance, the palmitoylation of signaling proteins can affect their interaction with membranes and other signaling molecules, thereby modulating cellular signaling pathways involved in tumor progression.
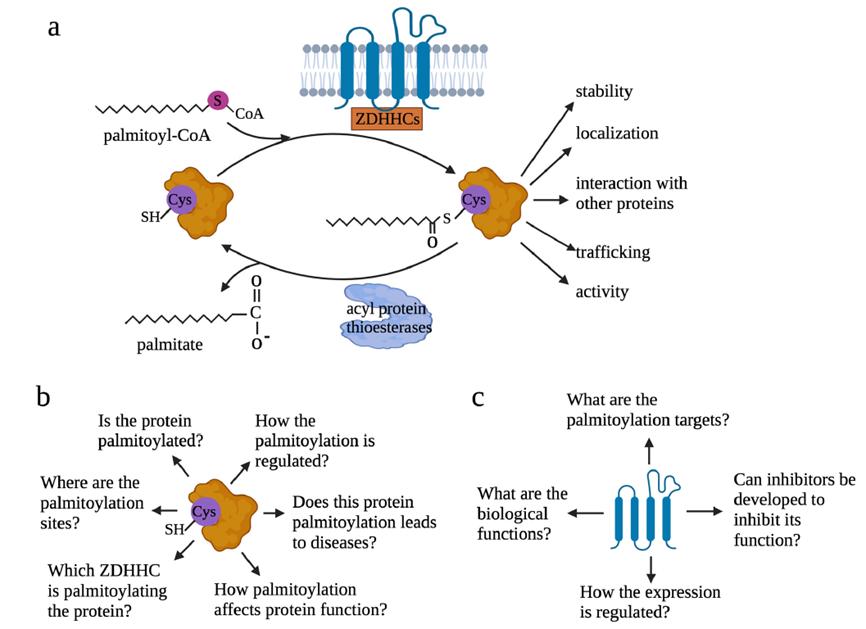 Figure 1: Overview of Palmitoylation and Key Issues
Figure 1: Overview of Palmitoylation and Key Issues
Key Issues in the Study of Protein Palmitoylation
Research Focus from the Perspective of Modified Proteins
The investigation of palmitoylation from the perspective of modified proteins necessitates addressing several core issues:
Identification of Palmitoylation: It is crucial to determine whether the target protein has undergone palmitoylation modification.
Localization of Palmitoylation Sites: Accurate identification of the specific cysteine residues that have undergone palmitoylation is essential.
Identification of ZDHHC Family Members: The specific ZDHHC family member(s) involved in the palmitoylation process must be identified.
Functional Impact Assessment: A comprehensive evaluation of how palmitoylation affects the function of the target protein is required.
Pathophysiological Role: It is important to investigate whether palmitoylation participates in and regulates specific pathophysiological processes.
Regulatory Mechanisms: An in-depth analysis of the regulatory mechanisms governing palmitoylation modifications should be conducted.
Research Focus from the Perspective of Palmitoylation Enzymes (ZDHHC)
When examining palmitoylation from the perspective of palmitoylation enzymes, specifically ZDHHC, the following key issues should be prioritized:
Role and Function of ZDHHC: The role and function of the ZDHHC enzyme in specific biological processes should be clearly defined.
Target Protein Identification: It is necessary to identify and determine the range of target proteins for specific ZDHHC enzymes.
Therapeutic Development Potential: The feasibility of developing novel therapeutic approaches by targeting specific ZDHHC enzymes should be explored.
Expression Patterns and Regulatory Mechanisms: The expression patterns of ZDHHC enzymes and the intrinsic regulatory mechanisms of their palmitoylation states should be thoroughly investigated.
Research Strategies for Palmitoylation
Overall, research on palmitoylation tends to follow a conventional framework, as illustrated in Figure 2.
 Figure 2: Research Approaches to Palmitoylation
Figure 2: Research Approaches to Palmitoylation
Analysis of Palmitoylation and Its Biological Significance
This article integrates insights from three studies: "DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis," "Palmitoylation-driven PHF2 ubiquitination remodels lipid metabolism through the SREBP1c axis in hepatocellular carcinoma," and "Palmitoylation-driven PHF2 ubiquitination remodels lipid metabolism through the SREBP1c axis in hepatocellular carcinoma." These studies elucidate methods for researching palmitoylation and its biological significance.
Determining Substrate Palmitoylation and Its Effects
DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis
A pivotal study published in Nature Communications in 2021 explored the palmitoylation of glucose transporter GLUT1 and its role in enhancing glycolysis and tumorigenesis in glioblastoma. GLUT1 is a key protein involved in glucose uptake, and its elevated expression is a critical molecular characteristic of cancer cells. GLUT1 is frequently overexpressed in various cancer types and is associated with poor prognosis. Consequently, GLUT1 remains a prominent focus in cancer research.
Experimental Approach and Findings
1. Impact of Palmitoylation on GLUT1 Localization
To investigate the effect of palmitoylation on GLUT1, the researchers employed a palmitoylation inhibitor, 2-BP, on glioblastoma cell lines. Immunofluorescence (IF) with GLUT1 antibodies was used to assess GLUT1's subcellular localization. The results revealed that 2-BP treatment led to a shift in GLUT1 localization from the plasma membrane (PM) to the cytoplasm in both cell lines (Figure 3a).
2. Western Blot Analysis
Soluble and membrane proteins from the cells were extracted and analyzed using Western blotting (WB). The analysis demonstrated that de-palmitoylated GLUT1, following 2-BP treatment, was localized to the cytosol, whereas palmitoylated GLUT1 was predominantly present on the plasma membrane (Figure 3b).
3. Detection of Palmitoylated GLUT1
Cells were labeled with a palmitate analog, alkynyl palmitate (Alkynyl PA), and probed with anti-palmitoylated GLUT1 antibodies via WB. The presence of palmitoylated GLUT1 was confirmed. Additionally, treatment with hydroxylamine (HAM), a palmitoylation-cleaving agent, rendered palmitoylated GLUT1 undetectable (Figure 3c).
4. Acyl-PEGyl Exchange Gel Shift Assay
The acyl-PEGyl exchange gel shift (APE) assay was employed to distinguish between palmitoylated and non-palmitoylated proteins based on their gel electrophoresis migration rates. The APE assay further validated the palmitoylation modification of GLUT1 in glioblastoma cell lines (Figure 3d).
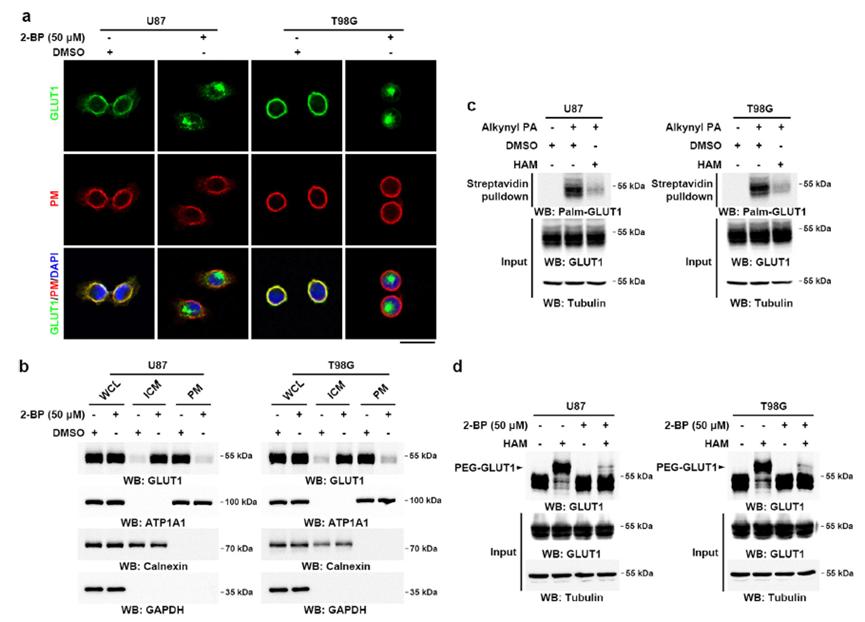 Figure 3: Acetylation Modification of GLUT1 Maintains Its Membrane Localization in GBM Cells
Figure 3: Acetylation Modification of GLUT1 Maintains Its Membrane Localization in GBM Cells
The aforementioned case study demonstrated how palmitoylation affects the subcellular localization of GLUT1. This was validated using IF and WB of different protein fractions. Reports have also highlighted that palmitoylation can influence the protein stability of PD-L1 and the ubiquitination activity of the E3 ubiquitin ligase PHF2. Therefore, to investigate the functional impact of palmitoylation on these proteins, distinct detection methods are required.
Palmitoylation Stabilizes PD-L1 to Promote Breast Tumor Growth
Palmitoylation influences various cellular processes, including protein stability and localization. A notable example is the stabilization of PD-L1 (Programmed Death-Ligand 1) through palmitoylation, which has implications for tumor growth and immune evasion in breast cancer.
Palmitoylation Verification
To validate palmitoylation of PD-L1, the study utilized antibodies specific for palmitoylated PD-L1. This approach allowed for the direct detection of the palmitoylated form of PD-L1. Additionally, the study employed Western blotting with PD-L1 antibodies to measure the overall protein levels of PD-L1, providing evidence of how palmitoylation impacts PD-L1 protein function and stability.
Detection of PD-L1 Protein Levels
The protein levels of PD-L1 were assessed using PD-L1-specific antibodies. This method offered insights into the functional consequences of palmitoylation on PD-L1, as changes in protein levels can reflect alterations in stability and activity induced by palmitoylation.
The study demonstrated that palmitoylation stabilizes PD-L1 protein levels, which contributes to enhanced tumor growth in breast cancer. The use of palmitoylation-specific antibodies confirmed the modification of PD-L1, while Western blotting revealed that palmitoylation affects PD-L1's stability and functional role within the tumor microenvironment (Figure 4).
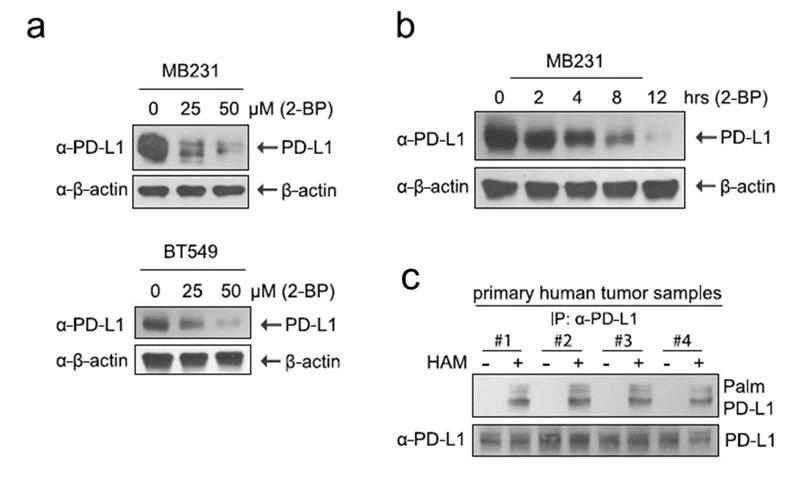 Figure 4: Palmitoylation Stabilizes PD-L1 Protein in Breast Cancer Cell Lines
Figure 4: Palmitoylation Stabilizes PD-L1 Protein in Breast Cancer Cell Lines
Palmitoylation-driven PHF2 ubiquitinationremodels lipid metabolism through theSREBPlc axis in hepatocellular carcinoma
Palmitoylation can significantly influence protein function and interactions. This study explores the role of palmitoylation in the ubiquitination of PHF2 (PHD finger protein 2) and its impact on lipid metabolism via the SREBP1c (Sterol Regulatory Element-Binding Protein 1c) axis in hepatocellular carcinoma (HCC).
Identification of Palmitoylated PHF2
Initial investigations confirmed that PHF2 undergoes palmitoylation. Following this, the focus shifted to understanding the functional implications of this modification. As PHF2 is known to function as an E3 ubiquitin ligase, its potential ubiquitination targets were explored.
Co-Immunoprecipitation (Co-IP) Assays
To identify and confirm the interaction between PHF2 and its ubiquitination substrate, SREBP1c, Co-IP assays were conducted. These assays demonstrated a direct interaction between PHF2 and SREBP1c (Figure 5a).
Subcellular Localization
The subcellular localization of PHF2 and SREBP1c was examined using fluorescence microscopy. The results showed that PHF2 and SREBP1c co-localize within the cells, further supporting their interaction (Figure 5b).
In Vitro Ubiquitination Assays
To validate the ubiquitination of SREBP1c by PHF2, in vitro ubiquitination assays were performed. The data confirmed that PHF2 catalyzes the ubiquitination of SREBP1c (Figure 5c).
The study elucidated the role of palmitoylation in regulating PHF2's function as an E3 ligase. It was established that PHF2, when palmitoylated, ubiquitinates SREBP1c, leading to alterations in lipid metabolism pathways. The interaction between PHF2 and SREBP1c, as well as their co-localization and the subsequent ubiquitination of SREBP1c by PHF2, were clearly demonstrated through the employed experimental techniques.
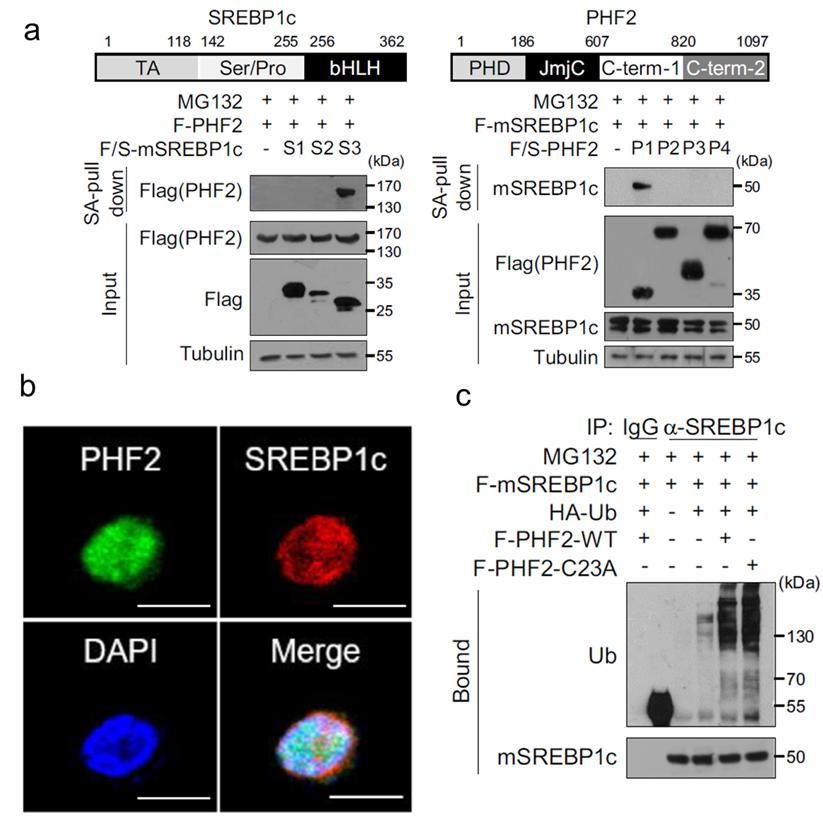 Figure 5: Palmitoylation Mediates Ubiquitination of SREBP1c by PHF2 in Hepatocellular Carcinoma
Figure 5: Palmitoylation Mediates Ubiquitination of SREBP1c by PHF2 in Hepatocellular Carcinoma
Determining Palmitoylation Sites in Substrates
Palmitoylation primarily occurrs on cysteine residues within proteins. Understanding the specific sites of palmitoylation is crucial for elucidating the modification's impact on protein function and cellular processes.
Site-Specific Mutation Analysis
A prevalent method for identifying palmitoylation sites involves site-specific mutagenesis of cysteine residues. This technique is valuable due to the limited number of potential palmitoylation sites, as palmitoylation exclusively occurs on cysteine residues.
For example, in the study of GLUT1, a glucose transporter protein, site-specific mutagenesis was employed. Each cysteine residue in GLUT1 was mutated, and subsequent analyses were performed to determine the palmitoylation status and localization of the protein.
In a recent study, site-specific mutagenesis was employed to determine the palmitoylation sites of GLUT1. Each cysteine residue was mutated, and the resulting proteins were analyzed for palmitoylation and localization. The results revealed that Cys207 is a critical palmitoylation site for GLUT1 (Figure 6).
Identifying palmitoylation sites is essential for understanding the functional consequences of this modification. Site-specific mutagenesis provides a powerful tool for pinpointing palmitoylation sites and elucidating their roles in protein function and cellular processes. The case of GLUT1 illustrates the utility of this approach in revealing critical modification sites and their impact on protein behavior.
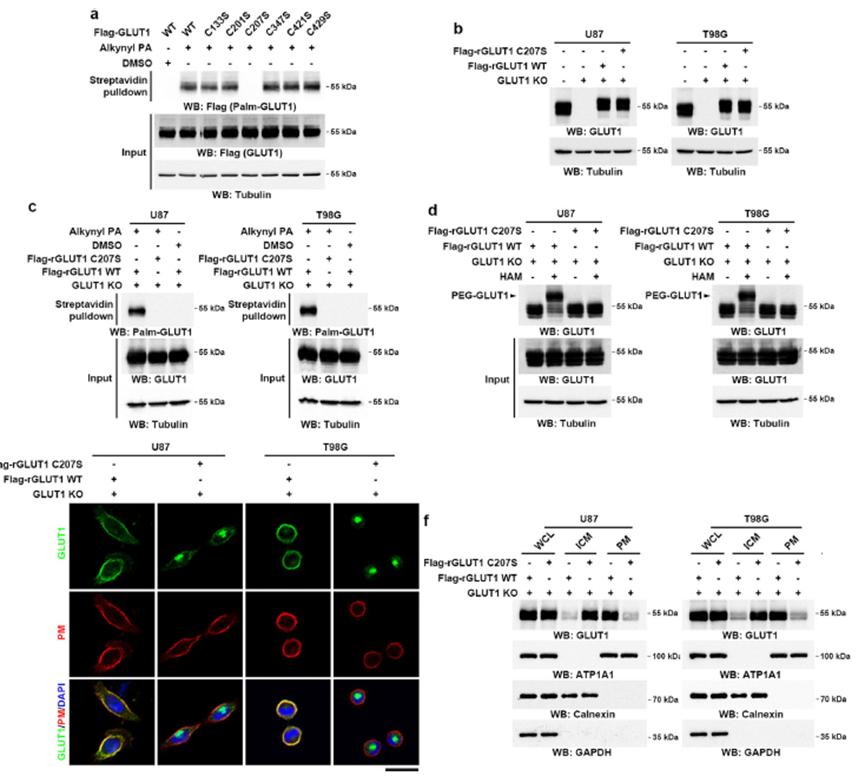 Figure 6: Cys207 as a Palmitoylation Site on GLUT1
Figure 6: Cys207 as a Palmitoylation Site on GLUT1
Database Utilization for Site Identification
A prominent method for identifying palmitoylation sites is through the use of dedicated palmitoylation prediction databases. These databases compile extensive information on known palmitoylation sites and utilize various algorithms to predict potential modification sites in target proteins.
In this study, the CSSPalm database (available at csspalm.biocuckoo.org) was employed to predict palmitoylation sites in the PD-L1 protein. The database utilizes comprehensive data and sophisticated algorithms to provide accurate predictions of palmitoylation sites based on sequence and structural information.
The analysis using CSSPalm identified Cys272 as a predicted palmitoylation site in PD-L1 (Figure 7). This result aligns with experimental findings and supports the utility of computational tools in identifying critical modification sites.
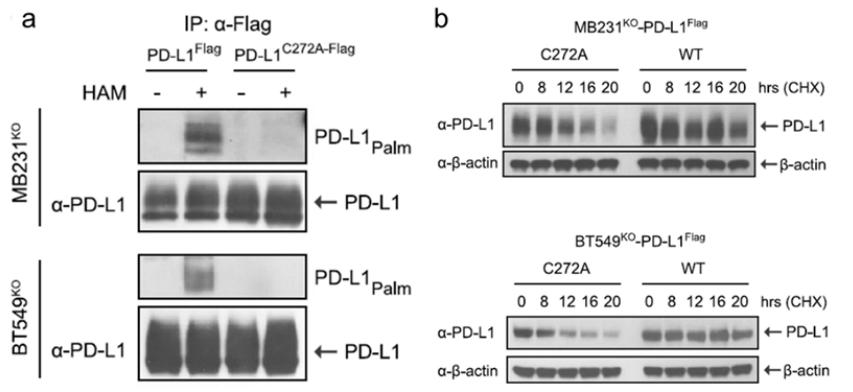 Figure 7: Cys272 as a Palmitoylation Site on PD-L1
Figure 7: Cys272 as a Palmitoylation Site on PD-L1
Identification of Palmitoylation Sites Using LC-MS
LC-MS provides a powerful approach for identifying palmitoylation sites by analyzing the molecular weight of modified peptides. The addition of palmitate, a lipid molecule, alters the mass of the amino acid residues to which it is attached. This change in molecular weight can be detected and quantified using mass spectrometry.
In this study, LC-MS was employed to identify palmitoylation sites on the PHF2 protein. The method involves the separation of peptides by liquid chromatography followed by mass spectrometric analysis to determine their molecular weights. This approach allows for the precise identification of modified residues based on their altered mass.
The LC-MS analysis identified Cys23 as a palmitoylation site on PHF2 (Figure 8). The mass shift observed in the spectra corresponded to the addition of palmitate, confirming the presence of palmitoylation at this specific residue.
The use of LC-MS for palmitoylation site identification offers high sensitivity and specificity, enabling the detection of even minor modifications. This method complements other techniques, providing a robust approach for validating and characterizing post-translational modifications.
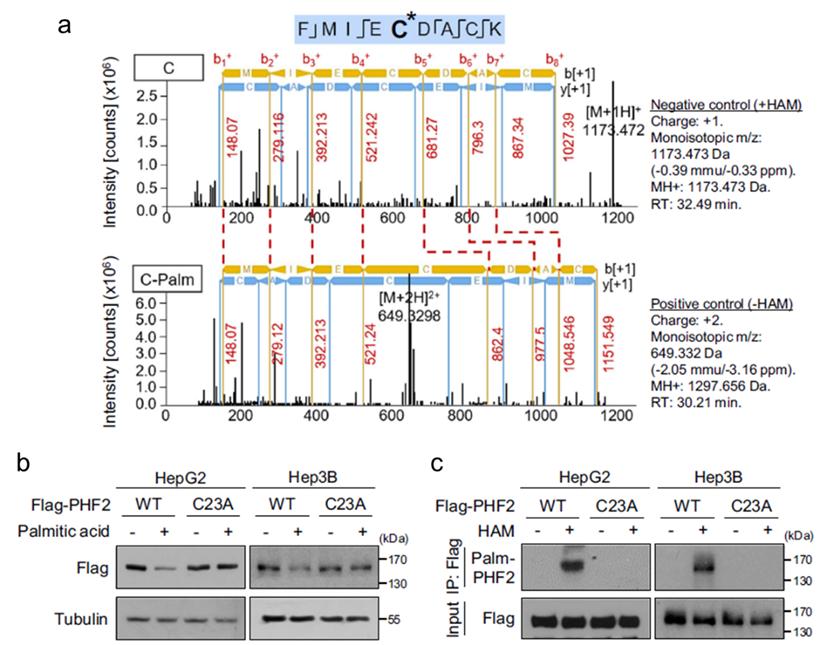 Figure 8: Cys23 as a Palmitoylation Site on PHF2
Figure 8: Cys23 as a Palmitoylation Site on PHF2
Screening and Identification of Palmitoylation Enzymes
Palmitoylation involves the covalent attachment of palmitic acid to cysteine residues in proteins. In mammals, a total of 23 palmitoylation enzymes are known. Identifying the specific enzymes responsible for the palmitoylation of a given substrate can be accomplished through targeted knockout or knockdown studies, as well as interaction studies using techniques such as Immunoprecipitation-Mass Spectrometry (IP-MS).
Identification of Palmitoylation Enzymes
To elucidate the palmitoylation enzyme responsible for modifying GLUT1, the following approach was utilized:
Gene Editing Using CRISPR/Cas9: Guide RNAs (gRNAs) targeting each of the 23 palmitoylation enzyme genes were designed and employed to generate knockout or knockdown cell lines. This technique facilitated the investigation of the impact of each enzyme on GLUT1 palmitoylation.
Analysis of GLUT1 Palmitoylation: Subsequent analysis of GLUT1 palmitoylation in these cell lines enabled the identification of DHHC9 as a crucial enzyme regulating GLUT1 palmitoylation. The palmitoylation status of GLUT1 was assessed in DHHC9 knockout cell lines, revealing a significant reduction in GLUT1 palmitoylation.
In Vitro Palmitoylation Assays: To confirm the role of DHHC9, in vitro palmitoylation experiments were conducted (Figures 9a, 9b). DHHC9 knockout and site-specific mutations were introduced to assess their effects on GLUT1 palmitoylation (Figures 9c, 9d). Further validation was performed using acylation protection assays (APE) (Figure 9e), which corroborated the involvement of DHHC9 in GLUT1 palmitoylation.
Subcellular Localization Analysis: Finally, the subcellular localization of GLUT1 was examined in DHHC9 knockout and mutant cell lines (Figures 9f, 9g). This analysis provided additional evidence supporting DHHC9 as the palmitoylation enzyme for GLUT1.
The combined results from gene editing, in vitro assays, and subcellular localization studies confirmed that DHHC9 is the specific palmitoylation enzyme responsible for modulating GLUT1. The reduction in GLUT1 palmitoylation upon DHHC9 knockout and the localization studies further solidify its role.
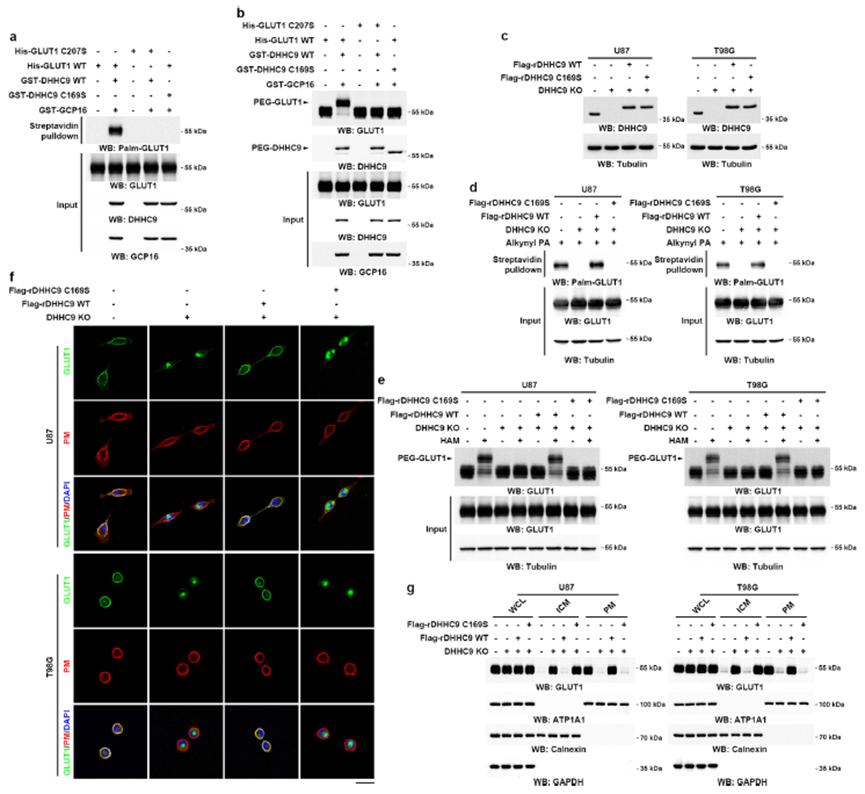 Figure 9: Palmitoylation of GLUT1 by DHHC9
Figure 9: Palmitoylation of GLUT1 by DHHC9
The enzyme ZDHHC9 was identified through Immunoprecipitation-Mass Spectrometry (IP-MS) as being involved in the palmitoylation of PD-L1. This method allowed for the specific detection of ZDHHC9's role in modifying PD-L1. The experimental approach and analytical techniques used to ascertain the involvement of ZDHHC9 in PD-L1 palmitoylation mirror those previously described for the palmitoylation of GLUT1 by DHHC9. Due to the methodological similarities, a detailed exposition of the experimental procedures used to identify ZDHHC9 as a palmitoylation enzyme for PD-L1 will not be provided in this discussion.
Biological Significance of Palmitoylation
Upon confirming that a specific protein undergoes palmitoylation, the subsequent critical inquiry pertains to the impact of this modification on cellular function and organismal physiology. The investigation can be approached from both cellular and animal model perspectives, with methodologies tailored to the functional role of the substrate protein.
Cellular Impact of Palmitoylation
For instance, GLUT1, a glucose transporter protein, requires specific research methods to elucidate its function. Studies on glycolysis, such as glucose uptake assays, extracellular acidification rate (ECAR) assays, and lactate production measurements, can elucidate the effects of GLUT1 palmitoylation on cellular function.
Similarly, PD-L1, an immune checkpoint molecule, necessitates experiments assessing T-cell cytotoxicity to analyze its functional implications. Further examination of tumor cell functionalities, including colony formation assays and proliferation assays such as MTT and CCK-8, can provide insights into how palmitoylation influences these cells.
In the case of PHF2, an E3 ligase, research on ubiquitination, including in vitro ubiquitination assays and treatment with MG132, can demonstrate the impact of palmitoylation on substrate function.
Animal Model Studies
At the animal model level, palmitoylation's regulatory role can be further elucidated by observing tumor size in xenograft mouse models and in mice with knockout of key proteins. Additional parameters such as animal growth and survival rates can provide further insights into the in vivo effects of palmitoylation.
Conclusion
Overall, while post-translational modifications such as palmitoylation may appear complex, detailed research methodologies simplify the experimental procedures and considerations. Palmitoylation research has gained widespread recognition due to its relatively straightforward research approaches and methodologies. However, as scientific advancements continue, a deeper exploration of the biological significance of palmitoylation is imperative. Future research trends will increasingly focus on extending investigations from cellular models to mouse models and clinical samples, with the aim of comprehensively elucidating its biological functions.
References
- Li et al., Diverse Roles of Protein Palmitoylation in Cancer Progression, Immunity, Stemness, and Beyond. Cells. 2023 Sep 5;12(18):2209.
- Zhang et al., DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis. Nat Commun. 2021 Oct 7;12(1):5872.
- Joost et al., Nomenclature of the GLUT/SLC2A family of sugar/polyol transport facilitators. Am J Physiol Endocrinol Metab. 2002
- Vander Heiden et al., Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009 May 22;324(5930):1029-33.
- Ancey et al., Glucose transporters in cancer—from tumor cells to the tumor microenvironment. FEBS J. 2018 Aug;285(16):2926-2943.
- Pouliot et al., Palmitoylation of the glucose transporter in blood brain barrier capillaries. Biochim Biophys Acta. 1995 Mar 22;1234(2):191-6.
- Yang et al., Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 2019 Jan;29(1):83-86.
- Jeong et al., Palmitoylation-driven PHF2 ubiquitination remodels lipid metabolism through the SREBP1c axis in hepatocellular carcinoma. Nat Commun. 2023 Oct 12;14(1):6370.
Our products and services are for research use only.


 Figure 1: Overview of Palmitoylation and Key Issues
Figure 1: Overview of Palmitoylation and Key Issues Figure 2: Research Approaches to Palmitoylation
Figure 2: Research Approaches to Palmitoylation Figure 3: Acetylation Modification of GLUT1 Maintains Its Membrane Localization in GBM Cells
Figure 3: Acetylation Modification of GLUT1 Maintains Its Membrane Localization in GBM Cells Figure 4: Palmitoylation Stabilizes PD-L1 Protein in Breast Cancer Cell Lines
Figure 4: Palmitoylation Stabilizes PD-L1 Protein in Breast Cancer Cell Lines Figure 5: Palmitoylation Mediates Ubiquitination of SREBP1c by PHF2 in Hepatocellular Carcinoma
Figure 5: Palmitoylation Mediates Ubiquitination of SREBP1c by PHF2 in Hepatocellular Carcinoma Figure 6: Cys207 as a Palmitoylation Site on GLUT1
Figure 6: Cys207 as a Palmitoylation Site on GLUT1 Figure 7: Cys272 as a Palmitoylation Site on PD-L1
Figure 7: Cys272 as a Palmitoylation Site on PD-L1 Figure 8: Cys23 as a Palmitoylation Site on PHF2
Figure 8: Cys23 as a Palmitoylation Site on PHF2 Figure 9: Palmitoylation of GLUT1 by DHHC9
Figure 9: Palmitoylation of GLUT1 by DHHC9