Glycosylation represents a ubiquitous post-translational modification mechanism whereby carbohydrate residues are appended to proteins and lipids, thereby exerting substantial influence over their conformational dynamics, molecular stability, and biological functionality. A nuanced comprehension of glycosylation sites assumes paramount importance in unraveling the multifaceted intricacies underlying the biology of glycoproteins and their intricate participation in a spectrum of physiological and pathological phenomena. The present discourse endeavors to undertake a comprehensive examination of methodologies and techniques employed in the elucidation of glycosylation sites, juxtaposing various mapping strategies, and delineating the distinctive attributes characterizing N-linked and O-linked glycosylation sites.
Introduction to Glycosylation Sites
Glycosylation sites serve as loci within protein or lipid sequences where glycans undergo covalent attachment to specific amino acid residues. These sites assume pivotal functions in mediating protein folding, stability, intracellular trafficking, and molecular recognition processes. N-linked and O-linked glycosylation emerge as the predominant types, characterized by distinct enzymatic pathways and preferences for site occupancy.
N-Linked Glycosylation Sites
N-linked glycosylation occurs at specific consensus sequence motifs, notably Asn-X-Ser/Thr (where X represents any amino acid except proline), predominantly situated within the extracellular domains of membrane-bound and secreted proteins. Oligosaccharyltransferase, an enzyme localized within the endoplasmic reticulum (ER), mediates the attachment of glycans to asparagine residues at these sites. Notably, these sites exhibit considerable structural conservation and frequently play crucial roles in protein folding and quality control mechanisms.
O-Linked Glycosylation Sites
O-linked glycosylation involves the covalent attachment of glycans to serine or threonine residues located within unstructured regions of proteins. Unlike their N-linked counterparts, the identification of O-linked glycosylation sites poses challenges due to the absence of a stringent consensus sequence motif. This process is orchestrated by a diverse array of glycosyltransferases and glycosidases, primarily operating within the Golgi apparatus.
Glycosylation Sites Mapping Methods
Glycosylation, a sophisticated post-translational modification, assumes a pivotal role in sculpting protein structure, function, and molecular recognition. The delineation of glycosylation sites represents a fundamental pursuit aimed at elucidating the functional implications of glycosylation across a broad spectrum of biological processes. Within this discourse, we embark on an exploration of various methodologies and techniques deployed in the intricate endeavor of mapping glycosylation sites, encompassing traditional approaches as well as cutting-edge methodologies rooted in mass spectrometry.
Lectin Affinity Chromatography
Lectin affinity chromatography stands as a venerable technique utilized for the enrichment of glycoproteins from complex biological matrices, predicated upon the selective interactions between lectins—carbohydrate-binding proteins—and diverse glycan moieties. Through the immobilization of lectins onto chromatographic matrices, the technique enables the discerning capture and purification of glycoproteins from crude biological extracts. This method serves as a robust preparative step preceding downstream analyses, including mass spectrometry-based glycoproteomics. Noteworthy is the versatility inherent to lectin affinity chromatography, which permits customization to target specific glycan structures or classes of glycoproteins.
Hydrazide Chemistry-Based Enrichment
Hydrazide chemistry-based enrichment stands as an efficacious methodology for elucidating glycosylation sites within glycoproteins. Central to this approach is the covalent conjugation of glycoproteins to solid supports engineered with hydrazide functional groups. Under mild acidic conditions, glycoproteins undergo oxidation, yielding reactive aldehyde moieties on their glycan appendages. Subsequent reaction of these aldehyde groups with the hydrazide-functionalized solid support culminates in the immobilization of glycoproteins onto the substrate. Concomitant washing steps effectively remove non-glycosylated proteins and extraneous contaminants, leaving behind glycoproteins firmly affixed to the solid matrix. Enzymatic cleavage liberates glycopeptides for subsequent mass spectrometric analysis, facilitating the identification and cartography of glycosylation sites. Renowned for its heightened specificity and selectivity towards glycoproteins, hydrazide chemistry-based enrichment emerges as an invaluable asset in the arsenal of techniques for glycosylation site mapping.
Enzymatic Digestion with PNGase F
Peptide-N-Glycosidase F (PNGase F) digestion stands as a widely embraced methodology for the discernment of N-linked glycosylation sites within proteins. PNGase F orchestrates the hydrolytic cleavage of the glycosidic linkage linking the asparagine residue to the N-linked glycan moiety, thereby liberating deglycosylated peptides. This enzymatic cleavage process selectively eliminates the carbohydrate component while preserving the integrity of the peptide backbone. Subsequent interrogation of the deglycosylated peptides via mass spectrometry facilitates the identification and cartography of N-linked glycosylation sites. Renowned for its robustness and reliability, PNGase F digestion represents a stalwart approach in the exploration of N-linked glycosylation sites across a myriad of biological specimens.
Mass Spectrometry-Based Glycoproteomics
Mass spectrometry-based glycoproteomics stands at the vanguard of glycosylation sites mapping methodologies, offering unmatched sensitivity, resolution, and throughput. Leveraging advanced mass spectrometry platforms in concert with innovative fragmentation techniques such as electron transfer dissociation (ETD) and collision-induced dissociation (CID), enables exhaustive analysis of glycopeptides. Within glycoproteomics workflows, enzymatic digestion yields peptides from glycoproteins, subsequently subject to enrichment through affinity chromatography or analogous strategies. Following enrichment, mass spectrometric analysis affords insights into both amino acid sequence and glycan composition of glycopeptides. Augmented by stable isotope labeling methodologies, quantitative assessments of glycosylation site occupancy and dynamics become feasible. The advent of mass spectrometry-based glycoproteomics has wrought a paradigm shift within the realm of glycoscience, facilitating the systematic exploration of glycosylation sites across a diverse array of biological contexts.
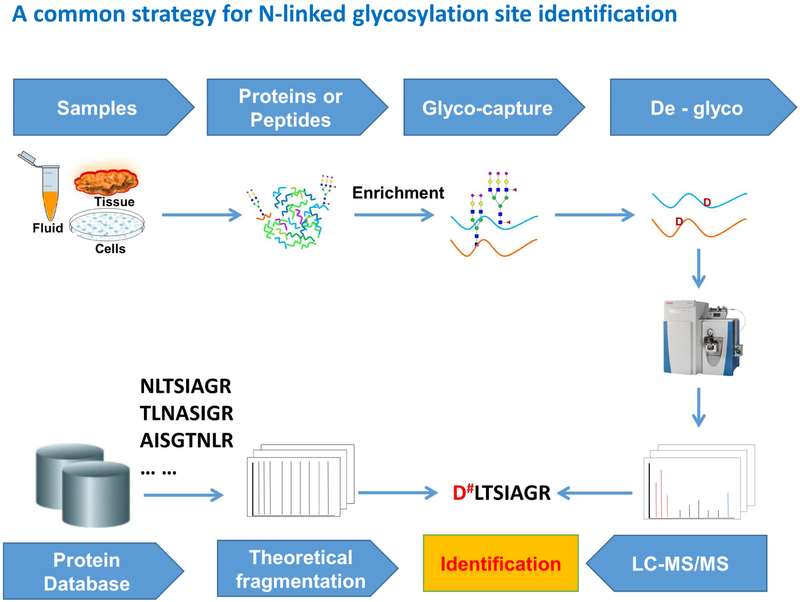 The most commonly used workflow for N-linked glycoprotein and glycosylation site identification.
The most commonly used workflow for N-linked glycoprotein and glycosylation site identification.
Comparison of Glycosylation Sites Mapping Methods
Traditional Versus High-Throughput Approaches
While traditional techniques like lectin affinity chromatography and enzymatic digestion utilizing PNGase F deliver robustness and reliability, they may fall short in addressing the throughput demands inherent in large-scale glycoproteomics investigations. Conversely, mass spectrometry-based glycoproteomics emerges as a potent alternative, boasting heightened sensitivity and throughput capabilities, thereby facilitating the concurrent examination of myriad glycosylation sites within complex biological matrices. Nonetheless, the adoption of mass spectrometry-based methodologies is impeded by the requisite specialized instrumentation and expertise, thereby constraining accessibility for certain researchers.
Specificity and Selectivity
Each method employed for mapping glycosylation sites manifests varying degrees of specificity and selectivity towards distinct glycan structures and classes of glycoproteins. Lectin affinity chromatography capitalizes on the binding specificity of lectins to glycan moieties, affording high specificity albeit limited selectivity towards particular glycan motifs. Conversely, enzymatic digestion utilizing PNGase F selectively targets N-linked glycosylation sites, thereby ensuring precise identification of such sites, albeit potentially overlooking O-linked glycosylation occurrences. In contrast, mass spectrometry-based glycoproteomics stands out for its unparalleled specificity and selectivity, facilitating the precise identification and quantification of glycosylation sites with remarkable precision and accuracy.
Advantages and Limitations
Each method employed for mapping glycosylation sites harbors its unique set of strengths and limitations. Lectin affinity chromatography offers simplicity and cost-effectiveness, yet it may encounter challenges related to nonspecific binding and restricted resolution capabilities. Enzymatic digestion utilizing PNGase F exhibits high specificity towards N-linked glycosylation sites; however, comprehensive glycosylation analysis may necessitate additional procedural steps. In contrast, mass spectrometry-based glycoproteomics affords a thorough characterization of glycosylation sites; nevertheless, its implementation mandates specialized instrumentation and bioinformatics proficiency. As a result, a synergistic amalgamation of complementary methodologies is frequently adopted to achieve a comprehensive delineation of glycosylation sites within intricate biological specimens.
Comparison of N-linked and O-linked Glycosylation Sites Mapping
N-linked and O-linked glycosylation delineate discrete post-translational modification pathways, each imparting distinctive structural and functional ramifications. The mapping of these glycosylation sites necessitates tailored methodological strategies, owing to variations in glycan attachment sites and enzymatic processing mechanisms.
Summary of Comparison of N-linked and O-linked Glycosylation Sites Mapping
| Aspect |
N-linked Glycosylation Sites Mapping Method |
O-linked Glycosylation Sites Mapping Method |
| Attachment Site |
Asparagine residues within Asn-X-Ser/Thr consensus sequence |
Serine or threonine residues |
| Enzymatic Processing |
Enzymatic digestion with PNGase F |
Specialized enrichment techniques (e.g., lectin chromatography, β-elimination) |
| Sequence Motif |
Asn-X-Ser/Thr (X ≠ Pro) |
Lacks specific consensus sequence motif |
| Predictability |
Structurally conserved, predictable |
Less predictable due to lack of specific sequence motif |
| Identification Challenges |
Empirical methods required due to lack of strict motif |
Challenges in predicting and identifying sites |
| Functional Implications |
Protein folding, stability, trafficking |
Cell adhesion, signaling, extracellular matrix remodeling |
| Site-Specific Analysis |
Enzymatic digestion with mass spectrometry |
Empirical methods, such as lectin binding assays |
| Applications in Biotechnology |
Optimization of therapeutic proteins, disease biomarker discovery |
Understanding disease mechanisms, therapeutic target identification |
Glycosylation Mapping:
N-linked glycosylation involves the attachment of glycans to the nitrogen atom of asparagine residues within the consensus sequence Asn-X-Ser/Thr (where X is any amino acid except proline). This process is typically mapped using enzymatic digestion with PNGase F to release N-linked glycans, followed by mass spectrometry analysis of the deglycosylated peptides.
For example, a study by An et al. (2019) employed mass spectrometry-based proteomics to identify and quantify N-linked glycosylation sites in Alzheimer's disease brain tissues. The researchers utilized PNGase F digestion coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS) to characterize the glycosylation profiles of amyloid precursor protein (APP) isoforms, revealing disease-associated alterations in glycosylation patterns.
In contrast, O-linked glycosylation occurs through the attachment of glycans to the oxygen atom of serine or threonine residues. Mapping O-linked glycosylation sites often requires specialized enrichment techniques, such as lectin affinity chromatography or β-elimination, to isolate O-linked glycopeptides for mass spectrometry analysis.
For instance, a study by Tian et al. (2020) employed lectin affinity chromatography coupled with LC-MS/MS to identify O-linked glycosylation sites in breast cancer tissues. The researchers utilized wheat germ agglutinin (WGA) lectin chromatography to enrich O-linked glycopeptides, followed by mass spectrometry analysis to characterize the glycosylation profiles of cancer-related proteins, revealing potential biomarkers for diagnostic and therapeutic applications.
Site-Specific Analysis
The structural differences between N-linked and O-linked glycosylation also influence the site-specific analysis of glycosylation sites. N-linked glycosylation sites are generally more predictable and have specific sequence motifs (Asn-X-Ser/Thr) that facilitate their identification. Consequently, mapping N-linked glycosylation sites can be achieved with high confidence using enzymatic digestion coupled with mass spectrometry analysis.
In contrast, O-linked glycosylation sites lack specific consensus sequences, making their identification more challenging. O-linked glycosylation sites are often identified through empirical methods, such as lectin binding assays or site-directed mutagenesis studies, which may not provide comprehensive coverage of O-linked glycosylation sites in proteins.
Functional Implications
The differences in the structural characteristics and enzymatic processing of N-linked and O-linked glycosylation sites have functional implications in biological systems. N-linked glycosylation is typically associated with protein folding, stability, and trafficking in the endoplasmic reticulum and Golgi apparatus. Moreover, N-linked glycans play critical roles in protein-protein interactions, immune recognition, and cell signaling.
On the other hand, O-linked glycosylation is involved in diverse cellular processes, including cell adhesion, signaling, and extracellular matrix remodeling. The site-specific analysis of N-linked and O-linked glycosylation sites enables the elucidation of their functional roles in health and disease.
Conclusion
Mapping Glycosylation sites is essential for elucidating the structure, function, and regulation of glycoproteins in biological systems. Various methods and techniques, including lectin affinity chromatography, hydrazide chemistry-based enrichment, enzymatic digestion with PNGase F, and mass spectrometry-based glycoproteomics, are employed for the comprehensive analysis of glycosylation sites. Each method offers unique advantages and challenges, and researchers often integrate multiple approaches to achieve comprehensive mapping of glycosylation sites in diverse biological samples. By advancing our understanding of glycosylation sites, researchers can unravel the complex biology of glycoproteins and their roles in health and disease.
References
- An, H. J., et al. (2019). Quantitative glycomics: A combined analytical and bioinformatics approach. Journal of Proteome Research, 18(1), 252-263.
- Tian, Y., et al. (2020). Site-specific glycosylation analysis of human plasma glycoproteins for biomarker discovery in breast cancer. Clinical Proteomics, 17(1), 8.
- An, H. J., Froehlich, J. W., & Lebrilla, C. B. (2009). Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Current Opinion in Chemical Biology.
- Kaji, H. (2015). Methods for large-scale glycosylation site mapping of N-glycoproteins. In N. Taniguchi, T. Endo, G. Hart, P. Seeberger, & C. H. Wong (Eds.), Glycoscience: Biology and Medicine. Springer.
- Hong, Q., Ruhaak, L. R., Stroble, C., Parker, E., Huang, J., Maverakis, E., & Lebrilla, C. B. (2015). A method for comprehensive glycosite-mapping and direct quantitation of serum glycoproteins. Journal of Proteome Research.
- Sun, S., & Zhang, H. (2015). Identification and validation of atypical N-glycosylation sites. Analytical Chemistry, 87(24), 11948-11951.
- Cao, L., Diedrich, J., Ma, Y. et al. Global site-specific analysis of glycoprotein N-glycan processing. Nat Protoc
- Dang L, Jia L, Zhi Y, Li P, Zhao T, Zhu B, Lan R, Hu Y, Zhang H, Sun S. Mapping human N-linked glycoproteins and glycosylation sites using mass spectrometry. Trends Analyt Chem. 2019
Our products and services are for research use only.


 The most commonly used workflow for N-linked glycoprotein and glycosylation site identification.
The most commonly used workflow for N-linked glycoprotein and glycosylation site identification.