In perusing scholarly literature, one frequently encounters terms such as E3 ubiquitin ligases and 26S proteasome, which are often associated with the concept of protein degradation. Initially, one may find these concepts somewhat nebulous, evoking a sense of partial comprehension intertwined with lingering ambiguity. However, upon delving deeper into the subject matter, it becomes apparent that these terms harbor numerous intricacies. Today, in collaboration with CD Genomics, we endeavor to unveil the veiled mysteries. Through the scrutiny and compilation of data, it is discerned that the aforementioned terms actually denote the ubiquitin-regulated pathways of protein degradation. Therefore, it is pertinent herein to furnish a succinct overview of pertinent background knowledge.
What is Ubiquitination
Ubiquitination is one of the post-translational modifications of proteins, and the interaction between the ubiquitin-proteasome system (UPS) and substrates forms a complex ubiquitination signaling network, participating in the regulation of almost all biological activities within organisms, including cell cycle progression, immune responses, and signal transduction. In-depth exploration of protein ubiquitination is of significant importance for elucidating the molecular mechanisms underlying its involvement in plant immunity, growth and development, and response to abiotic stress.
Ubiquitin and Ubiquitin Chains
Ubiquitin (Ub), a small protein molecule weighing approximately 8.5 kDa, comprises 76 amino acids and exhibits a high degree of evolutionary conservation. Within its full-length structure, ubiquitin harbors seven lysine (Lys) residues (designated as K6, K11, K27, K33, K48, and K63), one N-terminal methionine (Met) residue (M1), and one C-terminal glycine (Gly) residue (G76). Ubiquitin molecules bind via these specific residues, thereby forming ubiquitin chains. If ubiquitin molecules are linked in the same manner, homotypic ubiquitin chains are formed; conversely, if linked differently, heterotypic ubiquitin chains are formed. Ubiquitin chains can adopt linear or branched configurations. Additionally, ubiquitin itself undergoes various modification processes.
For illustrative purposes, a graphical representation is provided. The left panel depicts the structure of a ubiquitin molecule, while the middle and right panels respectively illustrate schematic diagrams of branched heterotypic ubiquitin chains and linear heterotypic ubiquitin chains. From the diagram, it is evident that the linkage pattern of each ubiquitin molecule is not identical, thus classifying them within the realm of heterotypic ubiquitin chains.
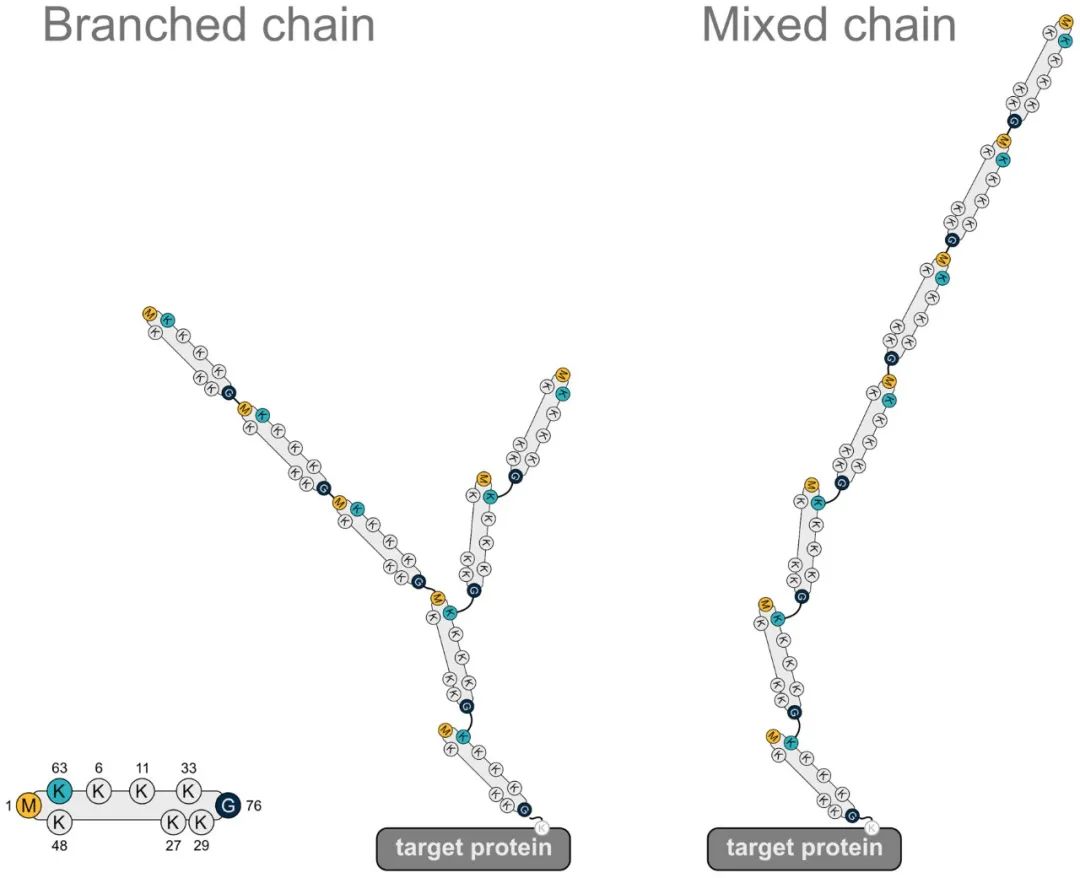 Figure 1 depicts a heterotypic ubiquitin chain containing the M1 linkage (Gunnar Dittmar and Konstanze F. Winklhofer, 2020).
Figure 1 depicts a heterotypic ubiquitin chain containing the M1 linkage (Gunnar Dittmar and Konstanze F. Winklhofer, 2020).
Note: The leftmost panel illustrates a ubiquitin molecule, the middle panel represents a heterotypic ubiquitin chain with branched structure, and the right panel shows a linear heterotypic ubiquitin chain.
Ubiquitination Pathway
Ubiquitination refers to the process by which ubiquitin is covalently attached to target proteins through the catalytic action of a series of enzymes. This series of enzymes encompasses three types that act in concert during the ubiquitination cascade: E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin-ligase enzyme.
The E3 ubiquitin ligases are primarily divided into two major families: the HECT (Homologous to the E6-AP Carboxyl Terminus) domain family and the RING (Really Interesting New Gene) domain family. Recently, a novel family of E3 ligases, the U-box protein family, has been identified. The HECT domain operates by forming a catalytic thioester bond with ubiquitin, which is essential for ubiquitin transfer. In contrast, the RING domain provides a docking site for the E2 enzyme and the substrate, thereby facilitating the E2-mediated transfer of ubiquitin to the substrate.
The Ubiquitination pathway proceeds through a series of enzymatic reactions, as illustrated in the accompanying figure. Initially, the E1 ubiquitin-activating enzyme utilizes ATP to activate ubiquitin, resulting in the formation of a ubiquitin-E1 (Ub-E1) complex. The ubiquitin moiety is then transferred from E1 to the E2 ubiquitin-conjugating enzyme through a transesterification reaction, forming a ubiquitin-E2 (Ub-E2) complex.
The ubiquitin moiety from the Ub-E2 complex can be transferred to the target protein via two distinct mechanisms. In the first mechanism, the E3 ubiquitin ligase directly facilitates the transfer of ubiquitin's C-terminal glycine to the ε-amino group of a lysine residue on the target protein. In the second mechanism, ubiquitin is initially transferred to the E3 ligase through transesterification, and subsequently, the E3 ligase recognizes the target protein and facilitates the transfer of ubiquitin's C-terminal glycine to the lysine residue of the target protein (Stone, 2019).
In summary, the ubiquitin molecule is covalently attached to target proteins or pre-existing ubiquitin chains on target proteins through the sequential catalytic actions of E1, E2, and E3 enzymes. The E3 ubiquitin ligase plays a crucial role in determining the specificity of target protein recognition. Furthermore, ubiquitination can also occur at the N-terminus of proteins and on other amino acids such as cysteine, serine, and threonine (Dittmar and Winklhofer, 2020; McClellan et al., 2019).
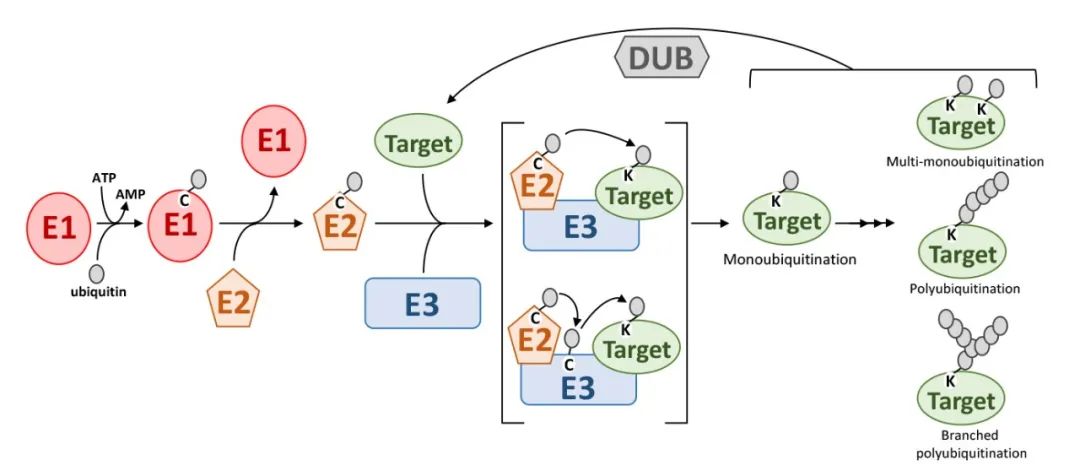 Figure 2. Ubiquitination Process (Stone, Sophia L., 2019) illustrates the ubiquitination process. The following abbreviations are used in the figure: E1 denotes the ubiquitin-activating enzyme; E2 represents the ubiquitin-conjugating enzyme; E3 signifies the ubiquitin-ligase enzyme; Target indicates the substrate protein; Ubiquitin refers to the ubiquitin molecule; and DUBs represent deubiquitinating enzymes.
Figure 2. Ubiquitination Process (Stone, Sophia L., 2019) illustrates the ubiquitination process. The following abbreviations are used in the figure: E1 denotes the ubiquitin-activating enzyme; E2 represents the ubiquitin-conjugating enzyme; E3 signifies the ubiquitin-ligase enzyme; Target indicates the substrate protein; Ubiquitin refers to the ubiquitin molecule; and DUBs represent deubiquitinating enzymes.
The attachment of a single ubiquitin molecule to the ε-amino group of a lysine residue on a target protein is referred to as monoubiquitination. When ubiquitin molecules are conjugated to multiple lysine residues on a target protein, the process is termed multiubiquitination. Polyubiquitination occurs when multiple ubiquitin molecules are attached to a single lysine residue on the target protein.
Deubiquitination
Ubiquitination is a tightly regulated and reversible process. Deubiquitinating enzymes (DUBs) can reverse ubiquitination by hydrolyzing the peptide or isopeptide bonds between ubiquitin molecules or between ubiquitin and substrate proteins (Fennell et al., 2018). Deubiquitinating enzymes not only inhibit the ubiquitination process but also promote it by degrading ubiquitination inhibitors, recycling ubiquitin molecules, and proofreading the ubiquitination process, thus forming a complex regulatory network with the ubiquitination system (Sowa et al., 2009).
Physiological Roles Associated with Different Types of Ubiquitin Chains
Based on the linkage sites of ubiquitin to substrates, there are currently nine recognized types of Ubiquitination: M1, K6, K11, K27, K29, K33, K48, K63, and G76. Each type of ubiquitination regulates distinct functions. Among these, K48-linked ubiquitination is associated with proteasomal degradation and is the most common, accounting for approximately 50% of all linkages. K6-linked ubiquitin chains may be involved in DNA damage responses and mitophagy. The K11/K48 branched chains enhance proteasomal degradation, with K11 ubiquitin chains also regulating the cell cycle. K27-linked ubiquitin chains serve as scaffolds for protein recruitment in the DNA damage response (DDR). K29-linked ubiquitination regulates lysosomal degradation. K33-linked ubiquitin chains are implicated in various biological processes. K63-linked ubiquitin chains and monoubiquitination may be involved in DNA repair and endocytosis. Linear ubiquitin chains linked via MET1 act as crucial positive regulators of NF-κB signaling, playing significant roles in tumorigenesis, inflammation, and immunity (Figure 3A).
Ubiquitination broadly participates in physiological processes such as immune response regulation, mitophagy, DNA damage repair, cell cycle control, epigenetic regulation, apoptosis, and protein degradation by modulating protein stability, subcellular localization, activity, and interactions (Figure 3A).
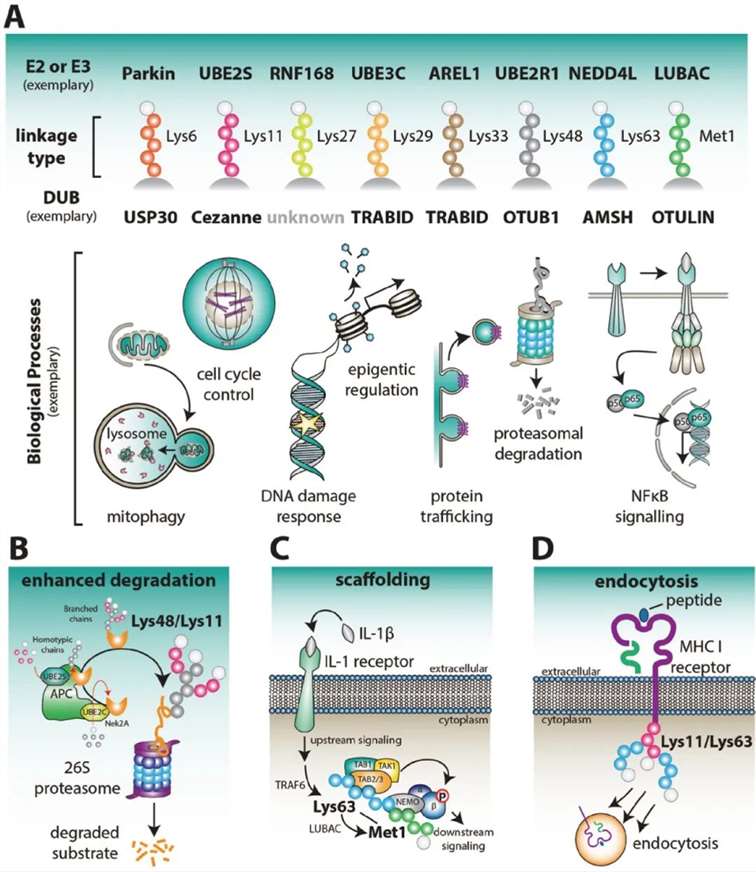 Figure 3: Physiological Functions Associated with Different Types of Ubiquitin Chains (Kirby N Swatek and David Komander, 2016)
Figure 3: Physiological Functions Associated with Different Types of Ubiquitin Chains (Kirby N Swatek and David Komander, 2016)
(A) Various Ubiquitin Chain Types and Associated Physiological Processes
Figure 3A illustrates the different types of ubiquitin chains and their involvement in typical physiological processes. Ubiquitin chains, depending on the linkage, play distinct roles in cellular functions.
(B) Role of APC/C in Early Mitosis
During early mitosis, the Anaphase-Promoting Complex/Cyclosome (APC/C) becomes active, regulating cell cycle factors such as Nek2A through Lys48/Lys11-linked polyubiquitination. In this process, UBE2C initially assembles short ubiquitin chains on the substrate, which are subsequently elongated by polymers linked via Lys11 on each ubiquitin. The Lys48/Lys11 branched chains enhance proteasomal degradation of these cell cycle regulators.
(C) Mixed or Branched Lys63/MET1 Chains in NF-κB Signaling
Mixed or branched Lys63/MET1 ubiquitin chains function as scaffolds for immune receptors, such as the interleukin-1 (IL-1) receptor, facilitating NF-κB signaling. These chains play a crucial role in propagating immune responses by enhancing signal transduction pathways.
(D) Viral E3 Ligases and MHC Class I Receptor Endocytosis
Viral E3 ligases initiate the endocytosis and internalization of major histocompatibility complex (MHC) class I receptors by attaching mixed or branched ubiquitin chains linked via Lys11/Lys63. This modification targets the receptors for degradation, thereby modulating immune evasion mechanisms employed by viruses.
Ubiquitin-proteasome system
Through extensive investigation, Ubiquitination has been identified as a critical regulator of various physiological functions within organisms. This study specifically focuses on the ubiquitin-mediated protein degradation pathway. Approximately 80% to 90% of intracellular proteins are degraded via the ubiquitin pathway, highlighting its significance in cellular metabolism (Pickart, Cecile M., and David Fushman, 2004).
The ubiquitin-controlled protein degradation process primarily involves several key steps. Initially, proteins targeted for degradation are tagged with ubiquitin, predominantly through K48-linked ubiquitin chains. It has been demonstrated that typically four K48-linked ubiquitin molecules are required for effective recognition by the proteasome. Although subsequent research has provided updates to this model, detailed discussion of these findings is beyond the scope of this paper; interested readers are encouraged to consult the relevant literature for further information.
Subsequently, the proteasome, principally the 26S proteasome, specifically recognizes and binds these ubiquitin-tagged substrate proteins, initiating the degradation process. This system, known as the ubiquitin-proteasome system (UPS), plays a crucial role in maintaining cellular homeostasis and regulating the cell growth cycle, DNA replication, and chromosomal architecture.
Therefore, ubiquitin-mediated protein degradation not only facilitates the removal of aberrant proteins but also exerts pivotal regulatory functions at multiple levels of cellular physiology.
Proteasome and 26S Proteasome
The proteasome, approximately 20S in size, exhibits a distinctive barrel-shaped structure and possesses the capability to degrade nearly all proteins into peptide chains of 7 to 9 amino acids in length. Its core functional region, the active site for protein degradation, is located at the center of the barrel-shaped structure, isolated from other cellular structures. The sole pathway for entry into this active site is facilitated by a complex known as the 19S regulator, which accurately recognizes ubiquitinated proteins, unfolds their structure, and assists the proteins in traversing the narrow channel of the proteasome to reach the active site located at the center of the barrel-shaped structure.
Once proteins are degraded into short peptides of 7 to 9 amino acids in length, these peptides are systematically released from the proteasome's opposite end. Additionally, noteworthy is the presence of a peptidase within the 19S regulator, which precisely removes ubiquitin from substrate proteins, ensuring that the removed ubiquitin can be recycled.
The 26S proteasome system, as a form of the proteasome, comprises two subcomplexes, the 20S proteasome, and the 19S proteasome, working together in highly coordinated mechanisms.
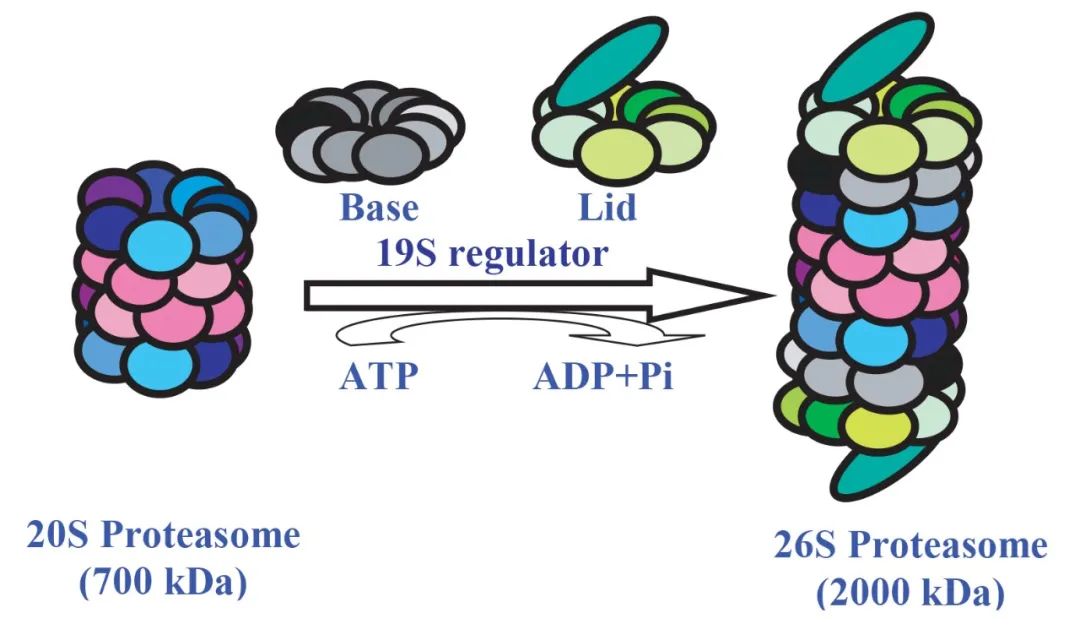 Figure 4 Formation of 26S proteasome, this process requires the consumption of ATP (Nandi et al., 2006).
Figure 4 Formation of 26S proteasome, this process requires the consumption of ATP (Nandi et al., 2006).
Prediction of Ubiquitination Sites
In the investigation of protein functions, if there is uncertainty regarding the Ubiquitination capacity of a protein, preliminary predictions can be conducted using bioinformatics tools, followed by targeted experimental validation. Several commonly utilized tools for predicting protein ubiquitination sites are recommended, as follows:
GPS-SUMO: Utilized for predicting SUMOylation sites and SUMO-interaction motifs (available at biocuckoo.cn).
ProP 1.0: A ubiquitination prediction service provided by DTU Health Tech-Bioinformatic Services.
GPS-Uber: A tool designed to predict the relationships between ubiquitin ligases and their substrates (available at biocuckoo.cn).
UbiNet 2.0: Accessible at cuhk.edu.cn.
CUCKOO Workgroup: Available at biocuckoo.org.
It is imperative to emphasize that these websites provide predictive functionality only. To ensure the accuracy and reliability of the results, experimental validation is required.
Detection of Protein Ubiquitination
The detection of protein ubiquitination is an essential technique for identifying ubiquitinated proteins, ubiquitination sites, and ubiquitination levels. Commonly employed methods include mass spectrometry (MS), immunoblotting, fluorescence polarization, and tandem affinity purification.
Mass Spectrometry
Mass spectrometry (MS) is a high-throughput technique utilized to identify and analyze protein ubiquitination. This method is capable not only of efficiently identifying ubiquitinated proteins but also of pinpointing specific ubiquitination sites and quantifying ubiquitination levels. Given that ubiquitinated proteins in vivo are typically prone to degradation and present in low abundance, it is necessary to enrich ubiquitinated peptides prior to MS analysis to increase their abundance. The general workflow encompasses five main steps: total protein extraction, proteolysis, enrichment of ubiquitinated peptides, MS analysis, and data interpretation.
In 2020, the journal Genomics Proteomics Bioinformatics published a research article entitled "Ubiquitinome Profiling Reveals the Landscape of Ubiquitination Regulation in Rice Young Panicles." In this study, the authors employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to analyze the ubiquitination status of proteins in rice (Oryza sativa) young panicles. The analysis identified 916 ubiquitinated proteins and 1,638 ubiquitination sites. Notably, 710 of the identified ubiquitinated proteins and 1,375 ubiquitination sites had not been previously reported (Figure 5).
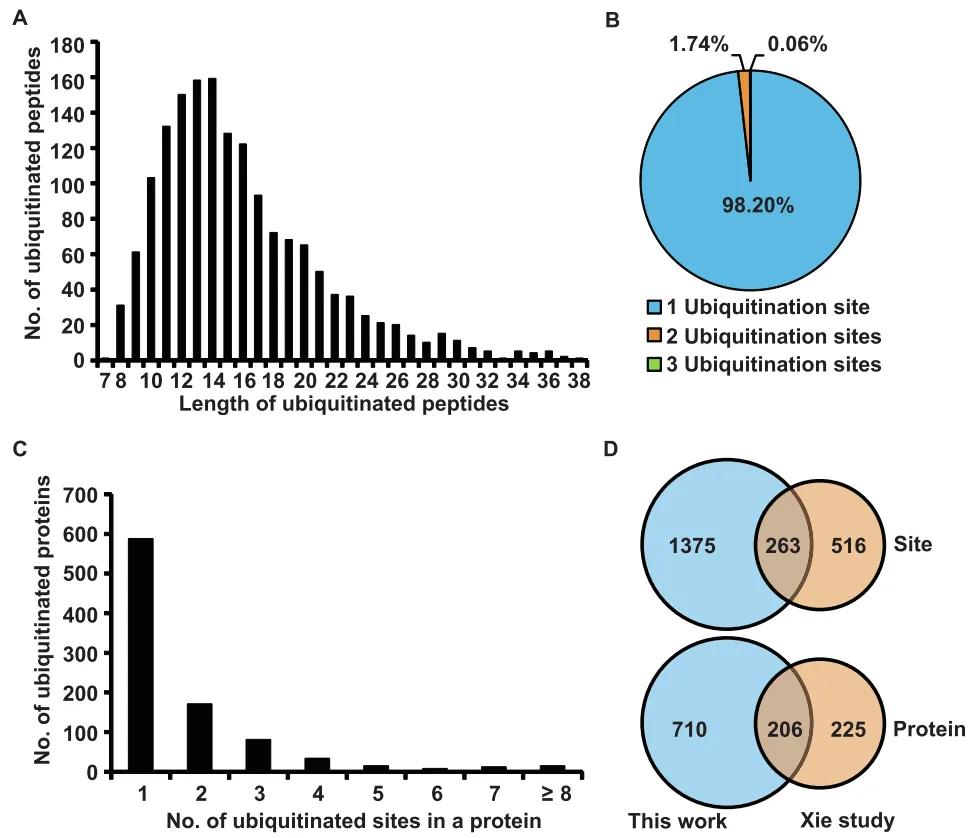 Figure 5: Characteristics of Identified Lysine-Ubiquitinated Peptides in Rice Young Panicles (Zhu et al., 2020)
Figure 5: Characteristics of Identified Lysine-Ubiquitinated Peptides in Rice Young Panicles (Zhu et al., 2020)
(A) Distribution of ubiquitinated peptides across different peptide lengths.
(B) Distribution of ubiquitinated peptides based on the number of ubiquitination sites.
(C) Distribution of ubiquitinated proteins based on the number of ubiquitination sites.
(D) Comparison of the number of ubiquitination sites and ubiquitinated proteins between young panicles and young leaves (Xin et al., 2015).
Immunoblotting
Immunoblotting is employed to detect protein ubiquitination and to analyze the expression levels and ubiquitination degrees of proteins under various physiological conditions. The fundamental principle involves the use of ubiquitination-specific antibodies that can recognize and bind to ubiquitinated proteins. Initially, proteins are extracted from cell or tissue samples and separated via SDS-PAGE electrophoresis. The separated proteins are then transferred onto a membrane and incubated with an anti-ubiquitination antibody. The presence of ubiquitinated proteins is indicated by higher molecular weight ubiquitinated bands. To detect the ubiquitination status of specific proteins, specific antibodies against the protein of interest can be utilized for immunoblotting.
In 2022, the journal Plant Biotechnology Journal published a research article entitled "Ubiquitin ligase OsRINGzf1 regulates drought resistance by controlling the turnover of OsPIP2;1." This paper elucidates the identification of the gene OsRINGzf1, which encodes an E3 ubiquitin ligase in rice (Oryza sativa), under drought conditions. Detailed domain analysis of the OsRINGzf1 protein was conducted, followed by successful in vitro expression and purification. Protein immunoblotting confirmed the E3 ubiquitin ligase activity of OsRINGzf1, as illustrated in Figure 6.
To investigate the interaction between OsRINGzf1 and OsPIP2;1, the authors employed co-immunoprecipitation (Co-IP) and split-luciferase complementation (Spilt-LUC) assays. To further ascertain whether OsPIP2;1 is a ubiquitination substrate of OsRINGzf1, a GFP-OsPIP2;1 fusion protein was constructed. Immunoblotting using anti-GFP and anti-ubiquitin antibodies revealed that OsPIP2;1 undergoes ubiquitination in vivo. In vitro ubiquitination assays corroborated that OsPIP2;1 is indeed a ubiquitination target of OsRINGzf1, as demonstrated in Figure 7.
This study provides novel insights into the mechanisms underlying rice drought stress responses. The identification of OsRINGzf1 and its regulatory role in the turnover of OsPIP2;1 offers potential genetic resources and theoretical foundations for breeding drought-resistant rice varieties.
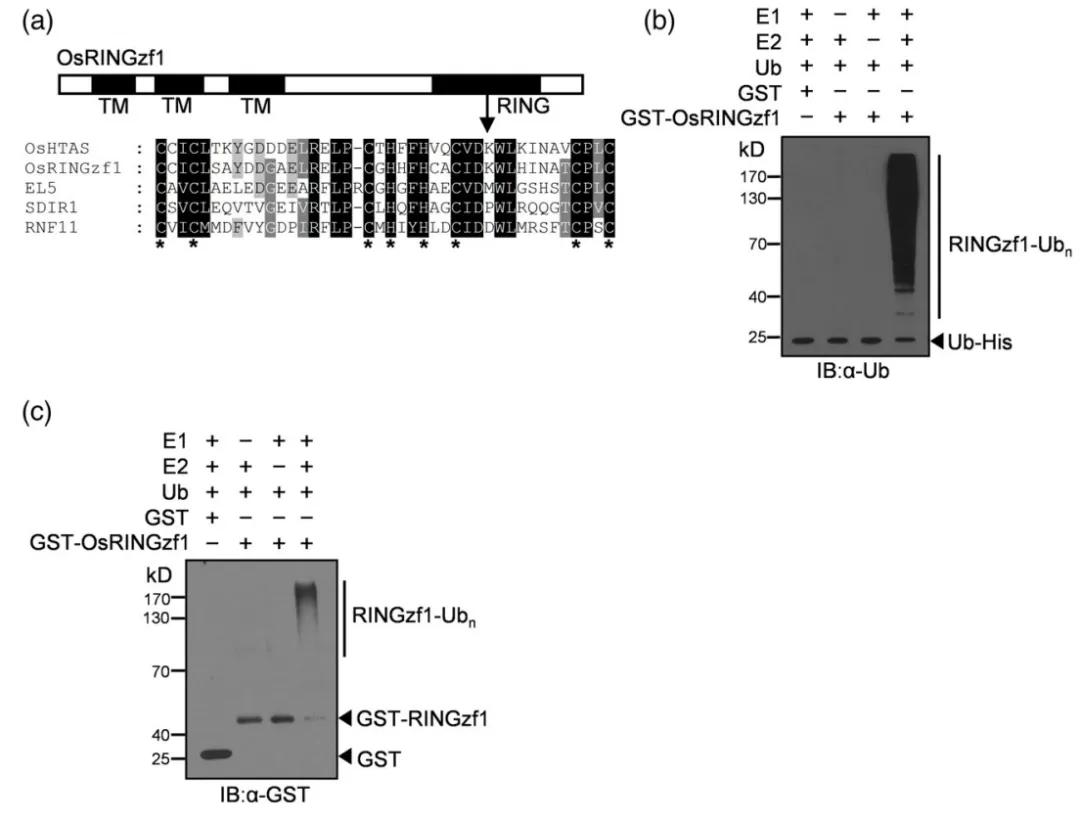 Figure 6: Ubiquitin Ligase Activity of OsRINGzf1 (Chen et al., 2022)
Figure 6: Ubiquitin Ligase Activity of OsRINGzf1 (Chen et al., 2022)
(a) OsRINGzf1 is characterized as a RING-H2 type E3 ligase.
(b-c) In the presence of E1, E2, and ubiquitin proteins, ubiquitination and GST antibodies were used to detect the autoubiquitination of OsRINGzf1-GST.
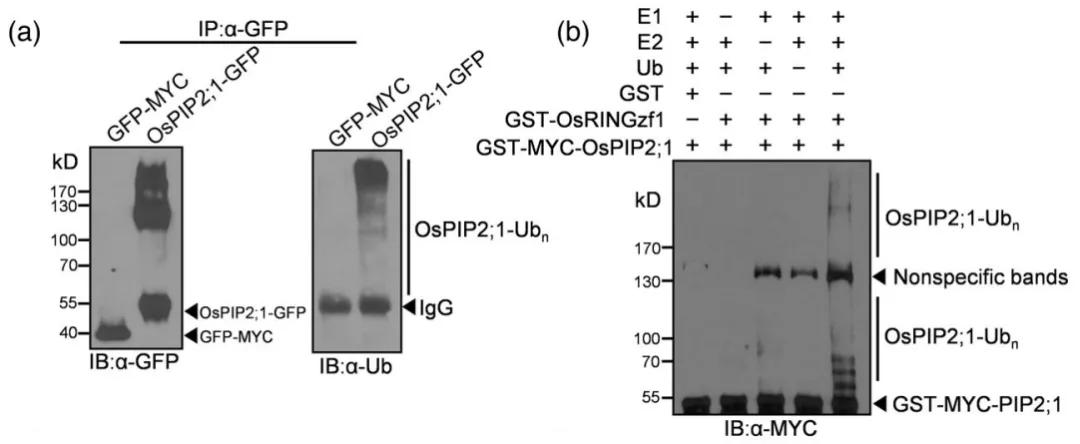 Figure 7: Ubiquitination of OsPIP2;1 Mediated by OsRINGzf1 (Chen et al., 2022)
Figure 7: Ubiquitination of OsPIP2;1 Mediated by OsRINGzf1 (Chen et al., 2022)
(a) In vivo ubiquitination of OsPIP2;1 protein, with GFP-MYC used as a negative control.
(b) In vitro ubiquitination of OsPIP2;1 protein, with GST serving as a negative control.
Fluorescence Polarization Assay
In 2019, the journal Frontiers in Chemistry published a research article titled "A high-throughput assay for monitoring ubiquitination in real time." In this study, the authors presented a novel approach for detecting protein ubiquitination, termed UbiReal. The fundamental principle of UbiReal involves fluorescent labeling of ubiquitin and real-time monitoring of the entire process of substrate protein ubiquitination using highly sensitive and stable fluorescence polarization technology. This technique enables the assessment of the activity of various enzymes involved in ubiquitination throughout the ubiquitination process and the detection of different types of ubiquitin modification (Figure 8).
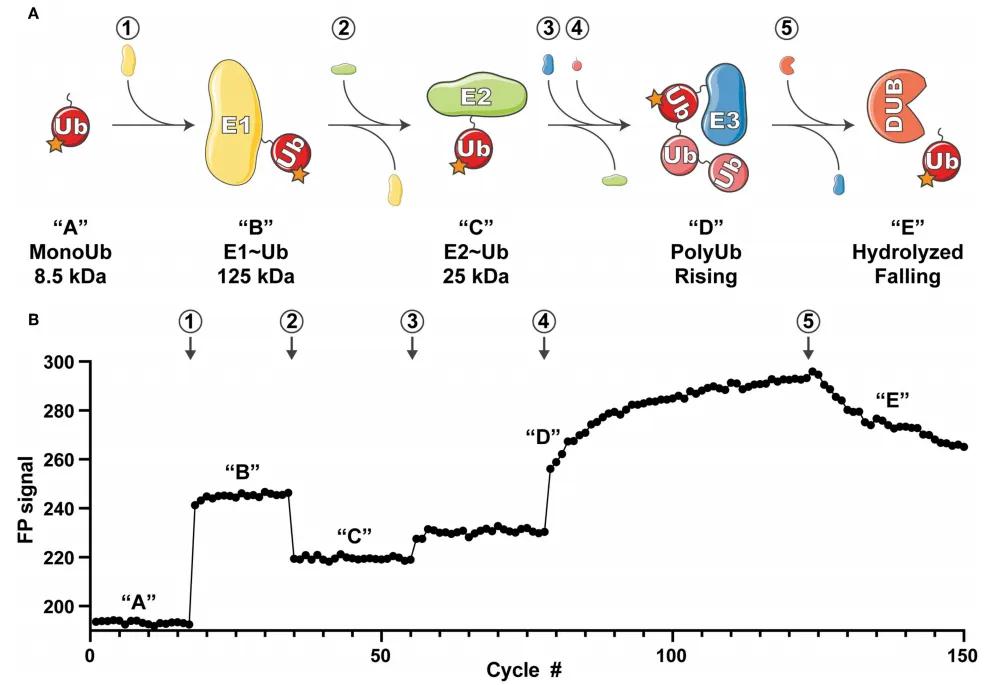 Figure 8: Overview of UbiReal Experimental Principle (Franklin and Pruneda, 2019)
Figure 8: Overview of UbiReal Experimental Principle (Franklin and Pruneda, 2019)
(A) Schematic representation of the ubiquitin conjugation sequence monitored by UbiReal, with numbers enclosed in circles indicating the addition of corresponding proteins. The letters in quotes represent the states of ubiquitin complexes, corresponding to the changes in fluorescence polarization (FP) signals observed in (B). Approximate molecular weights of complexes are provided, reflecting the expected amplitude of FP signals. "Rising" in (D) corresponds to the increase in FP signal generated by E3 enzyme activity, while "Falling" in (E) corresponds to the decrease in FP signal resulting from DUB enzyme activity.
(B) Data reflecting the comprehensive monitoring capability of UbiReal throughout the ubiquitination process. Numbers and letters denote the addition of corresponding proteins and the states of ubiquitin complexes, respectively.
Tandem Ubiquitin Binding Domains (UBD) Approach
This method is based on the principle that proteins containing ubiquitin binding domains (UBD) can recognize ubiquitination signals.
In 2019, the journal Analytical Chemistry published a research article titled "Specific and Unbiased Detection of Polyubiquitination via a Sensitive Non-Antibody Approach." In this study, the authors, building upon previous research, engineered tandem hybrid UBDs (ThUBD), which exhibited enhanced detection efficiency and affinity for ubiquitin chains compared to naturally occurring UBDs. ThUBD was employed in conjunction with Far-Western blotting to detect protein ubiquitination modifications (Figure 9), and this visualization technique for studying protein ubiquitination was termed TUF-WB. The basic workflow involves protein separation via SDS-PAGE, membrane transfer, blocking, incubation with ThUBD or glutathione transferase (GST), followed by incubation with anti-GST antibodies and subsequent detection through visualization.
To validate the feasibility of TUF-WB, the authors conducted immunoblot analysis experiments based on ThUBD using human embryonic kidney 293T cells and Escherichia coli cell lysates as experimental materials. The experimental results demonstrated the high specificity of TUF-WB in detecting ubiquitination signals (Figure 10).
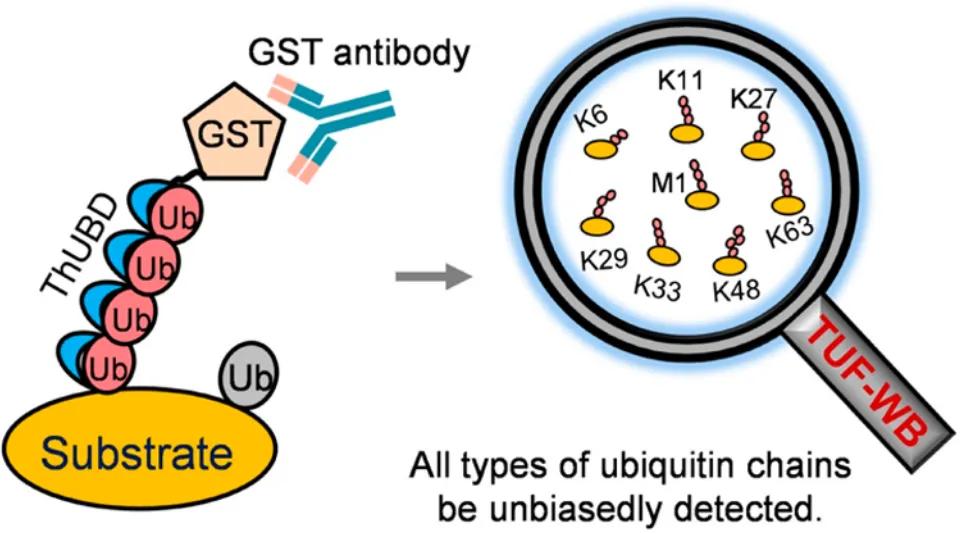 Figure 9 A new technology for detecting protein ubiquitination (TUF-WB) formed by ThUBD combined with Far-Western blot (Xiao et al., 2019).
Figure 9 A new technology for detecting protein ubiquitination (TUF-WB) formed by ThUBD combined with Far-Western blot (Xiao et al., 2019).
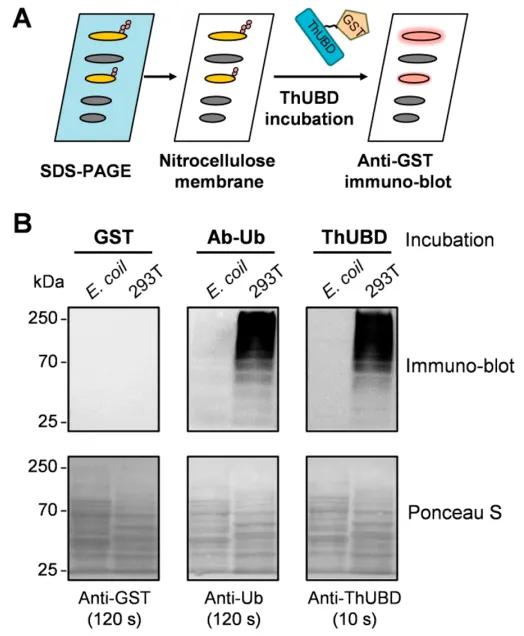 Figure 10: Specific Recognition of Ubiquitinated Proteins by TUF-WB (Xiao et al., 2019).
Figure 10: Specific Recognition of Ubiquitinated Proteins by TUF-WB (Xiao et al., 2019).
(A) The workflow of TUF-WB is delineated.
(B) Lysates from Escherichia coli and 293T cells immobilized on nitrocellulose membranes were incubated with GST antibody, ubiquitin antibody, and ThUBD. Detection with the anti-ubiquitin antibody served as a positive control. GST-tagged protein blotting was conducted, with the GST antibody detection serving as a negative control. Ponceau S staining was employed to verify protein loading.
The Role of Protein Ubiquitination in Plant Function
Involvement in Plant Immunity
Upon pathogenic invasion, plants employ physical barriers such as the cell wall and cuticle to initiate pattern-triggered immunity (PTI). Pathogens, in turn, secrete effector proteins to evade PTI, leading to the activation of effector-triggered immunity (ETI) as plants produce various resistance proteins that interact with these effectors. Post-translational modifications, including ubiquitination, regulate the signaling molecules and resistance proteins involved in PTI and ETI.
A study published in 2021 in Plant Physiology, entitled "Ubiquitylome Analysis Reveals a Central Role for the Ubiquitin-Proteasome System in Plant Innate Immunity," employed proteomics to identify ubiquitinated proteins in Arabidopsis thaliana following immune response activation. The study confirmed the involvement of the ubiquitin-proteasome system (UPS) in plant immunity.
The authors treated transgenic A. thaliana seedlings expressing hexa-6HisUBQ with exogenous flg22 (a conserved peptide from bacterial flagellin that induces innate immune responses). Ubiquitinated proteins were extracted from wild-type (WT), untreated control, and flg22-treated seedlings using a two-step affinity purification method (Figure 11). Mass spectrometry identified 690 high-confidence ubiquitinated proteins: 271 shared between control and flg22-treated groups, 299 unique to the flg22-treated group, and 120 unique to the control group. This indicates that flg22-triggered immune responses increase protein ubiquitination levels in A. thaliana (Figure 12A). Gene Ontology (GO) analysis revealed that the ubiquitinated proteins specific to the flg22-treated group are involved in multiple biological processes, including disease resistance and stress response (Figure 12B). Additionally, a significant presence of UPS-related proteins was observed among the ubiquitinated proteins (Figure 12C).
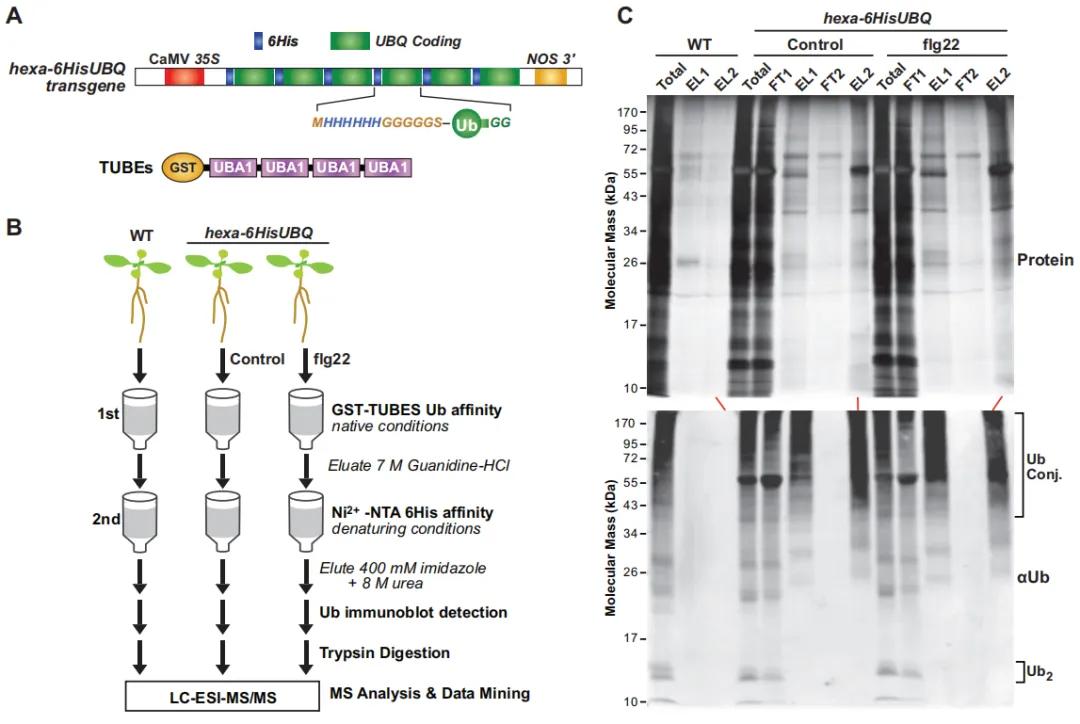 Figure 11: Affinity Purification of Ubiquitinated Proteins Expressing 6HIS-UBQ in Transgenic Arabidopsis thaliana (Ma et al., 2021).
Figure 11: Affinity Purification of Ubiquitinated Proteins Expressing 6HIS-UBQ in Transgenic Arabidopsis thaliana (Ma et al., 2021).
(A) Schematic representation of the hexa-6HIS-UBQ transgene. The synthetic UBQ gene encodes six tandem repeats of ubiquitin (Ub) monomers, with an N-terminal 6His tag followed by a glycine-rich linker (MHHHHHHGGGGGSA). The construct is driven by the constitutive CaMV 35S promoter, forming a single open reading frame encoding the hexa-6His-UBQ fusion protein (Saracco et al., 2009).
(B) Workflow diagram illustrating the proteomic analysis methodology for ubiquitinated proteins in Arabidopsis thaliana seedlings expressing p35S.
(C) Two-step affinity purification procedure used to isolate ubiquitinated proteins expressing 6His-Ub from Arabidopsis thaliana plants.
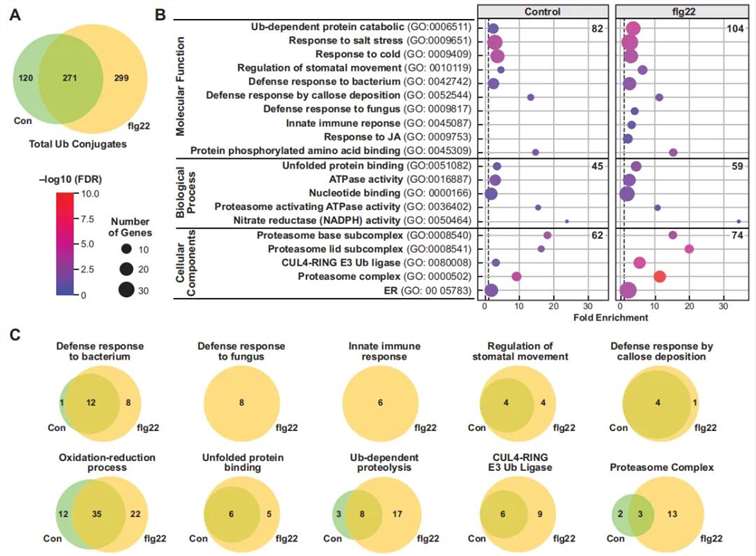 Figure 12: Gene Ontology (GO) Analysis of Ubiquitinated Proteins in flg22-Treated and Untreated Samples (Ma et al., 2021).
Figure 12: Gene Ontology (GO) Analysis of Ubiquitinated Proteins in flg22-Treated and Untreated Samples (Ma et al., 2021).
(A) Venn diagram illustrating the distribution of ubiquitinated proteins identified in control, flg22-treated, and untreated groups.
(B) GO annotation-based analysis of the biological processes, molecular functions, and cellular components associated with the ubiquitinated proteins.
(C) Functional grouping of proteins based on GO annotations, categorizing the proteins into distinct functional groups.
Previous studies have established that proteins associated with the ubiquitin-proteasome system (UPS) play a crucial role in plant immune responses. E3 ubiquitin ligases, which are ubiquitously present in the UPS, have been identified as key regulators of plant immunity (Zhang and Zeng, 2020). A recent study published in Horticulture Research in 2023, titled "A Putative E3 Ubiquitin Ligase Substrate Receptor Degrades Transcription Factor SmNAC to Enhance Bacterial Wilt Resistance in Eggplant," elucidates the molecular mechanism by which the substrate receptor protein SmDDA1b of an E3 ubiquitin ligase enhances resistance to bacterial wilt in eggplant through degradation of the transcription factor SmNAC. This study provides a theoretical foundation for genetic breeding in Solanaceae.
The authors employed a yeast two-hybrid screening assay to identify SmDDA1b as a potential interacting protein with SmNAC. Subsequent validation of the SmNAC-SmDDA1b interaction was performed using yeast two-hybrid (Y2H), bimolecular fluorescence complementation (BiFC), and co-immunoprecipitation (Co-IP) assays. Subcellular localization studies revealed that SmDDA1b is localized in the nucleus (Figure 13). Previous research indicated that SmNAC negatively regulates resistance to bacterial wilt in eggplant by inhibiting salicylic acid (SA) biosynthesis (Na et al., 2016).
To further investigate the role of SmDDA1b, the authors generated SmDDA1b overexpression and virus-induced gene silencing (VIGS) lines. Quantification of SA levels and expression analysis of SA biosynthesis-related genes (ICS1) and SA signaling pathway genes in these transgenic lines revealed that overexpression of SmDDA1b led to increased expression of these genes and elevated SA levels. Conversely, silencing of SmDDA1b resulted in decreased gene expression and SA levels. These findings indicate that SmDDA1b positively regulates SA biosynthesis and participates in its signaling pathway (Figure 14).
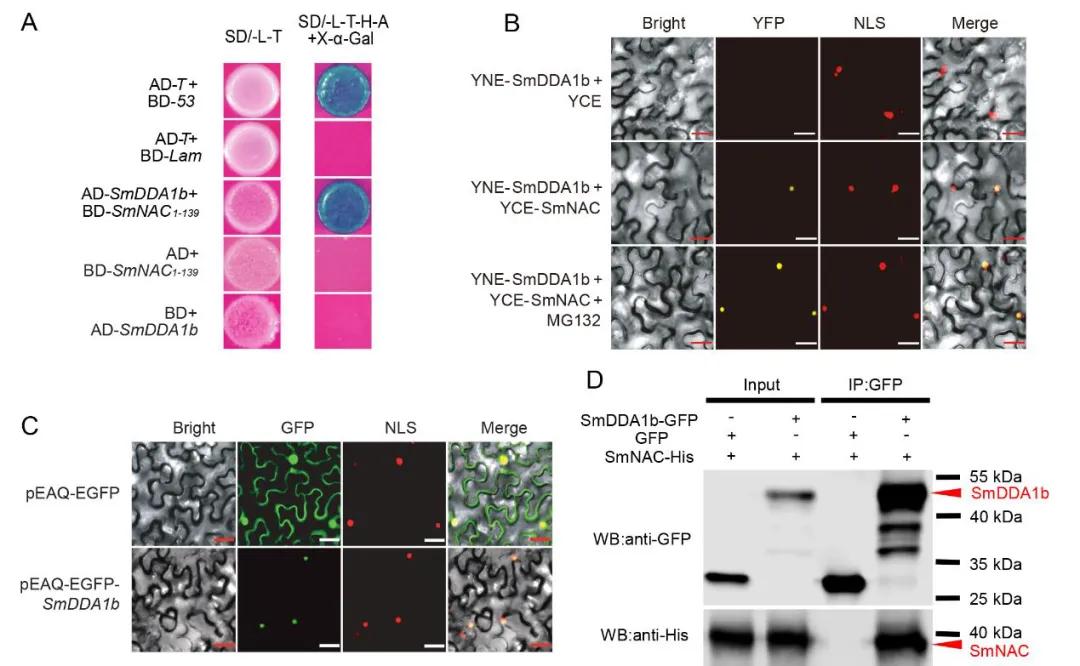 Figure 13: Interaction between SmDDA1b and SmNAC and Subcellular Localization of SmDDA1b (Yan et al., 2023).
Figure 13: Interaction between SmDDA1b and SmNAC and Subcellular Localization of SmDDA1b (Yan et al., 2023).
(A) Yeast two-hybrid (Y2H) assay demonstrating the interaction between SmDDA1b and SmNAC.
(B) Bimolecular fluorescence complementation (BiFC) assay confirming the interaction between SmDDA1b and SmNAC.
(C) Subcellular localization analysis showing the nuclear localization of SmDDA1b.
(D) Co-immunoprecipitation (Co-IP) assay further validating the interaction between SmDDA1b and SmNAC.
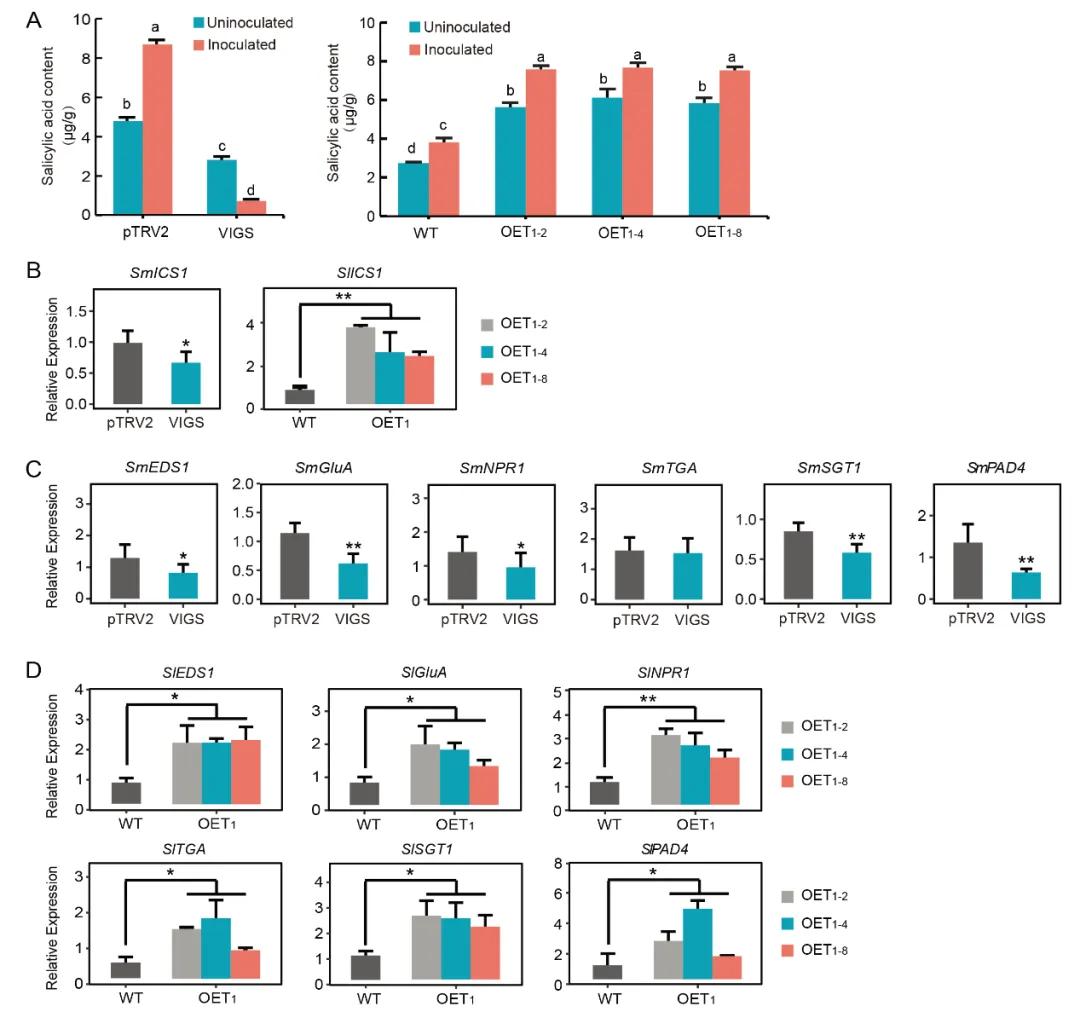 Figure 14: Positive Regulation of Salicylic Acid (SA) Biosynthesis and Participation in SA Signaling Pathways by SmDDA1b (Yan et al., 2023).
Figure 14: Positive Regulation of Salicylic Acid (SA) Biosynthesis and Participation in SA Signaling Pathways by SmDDA1b (Yan et al., 2023).
(A) Measurement of salicylic acid (SA) content in different Solanum melongena plant lines: pTRV2 (control group), VIGS (silenced plants), WT (wild-type), and OE (overexpression plants).
(B) Expression levels of ICS1 in silenced and overexpression plants.
(C, D) Expression levels of genes related to the SA signaling pathway in silenced and overexpression plants.
Studies have identified SmDDA1b as a homolog of AtDDA1, a known substrate receptor protein for the E3 ubiquitin ligase complex CRL4 (Irigoyen et al., 2014). The substrate receptor protein DDA1 interacts with the scaffold protein CUL4 of the cullin-RING ubiquitin-protein ligases (CRL4) through the DNA damage-binding protein DDB1 (Pang et al., 2019). Consequently, the authors employed yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays to verify the interactions between SmDDB1 and SmCUL4, as well as between SmDDB1 and SmDDA1b. These interactions suggest that SmDDA1b potentially exhibits E3 ubiquitin ligase activity in Solanum melongena (Figure 15A-D).
Subsequently, the authors conducted Western blot analysis, luciferase reporter assays, and subcellular localization studies to demonstrate that SmDDA1b mediates the ubiquitination and subsequent degradation of SmNAC (Figure 15E-F).
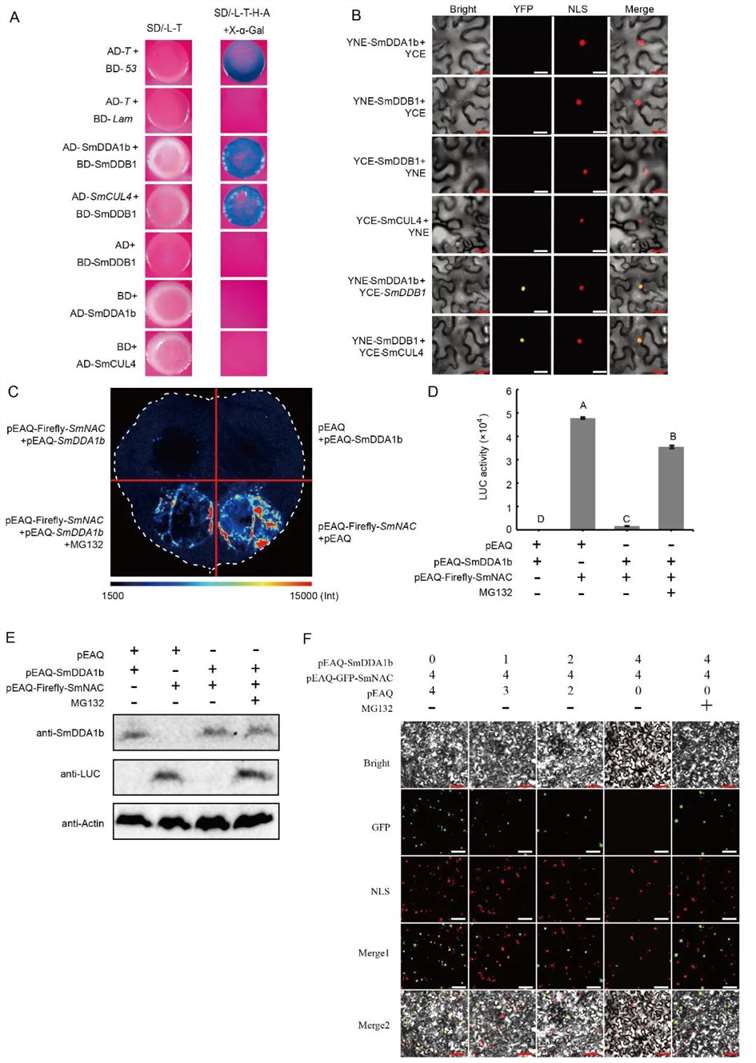 Figure 15: Interaction between SmDDB1, SmCUL4, and SmDDA1b, and SmDDA1b-Mediated Degradation of SmNAC (Yan et al., 2023).
Figure 15: Interaction between SmDDB1, SmCUL4, and SmDDA1b, and SmDDA1b-Mediated Degradation of SmNAC (Yan et al., 2023).
(A) Yeast two-hybrid (Y2H) assay demonstrating the interaction between SmDDB1 and SmCUL4.
(B) Bimolecular fluorescence complementation (BiFC) assay confirming the interaction between SmDDB1 and SmDDA1b.
(C) SmDDA1b-mediated degradation of SmNAC. pEAQ-Luciferase-SmNAC+pEAQ and pEAQ+pEAQ-SmDDA1b treatments serve as positive and negative controls, respectively, with MG132 as a proteasome inhibitor.
(D) Identification of luciferase activity.
(E) Western blot analysis.
(F) Subcellular localization experiment.
To further elucidate the defense mechanisms, the researchers investigated whether SmNAC could regulate SmDDA1b. Expression levels of SmDDA1b were assessed in SmNAC-overexpressing plants, revealing reduced expression compared to wild-type plants. Through yeast one-hybrid (Y1H) and Dual-LUC experiments, it was observed that SmNAC binds to the promoter of SmDDA1b and suppresses its expression (Figure 16).
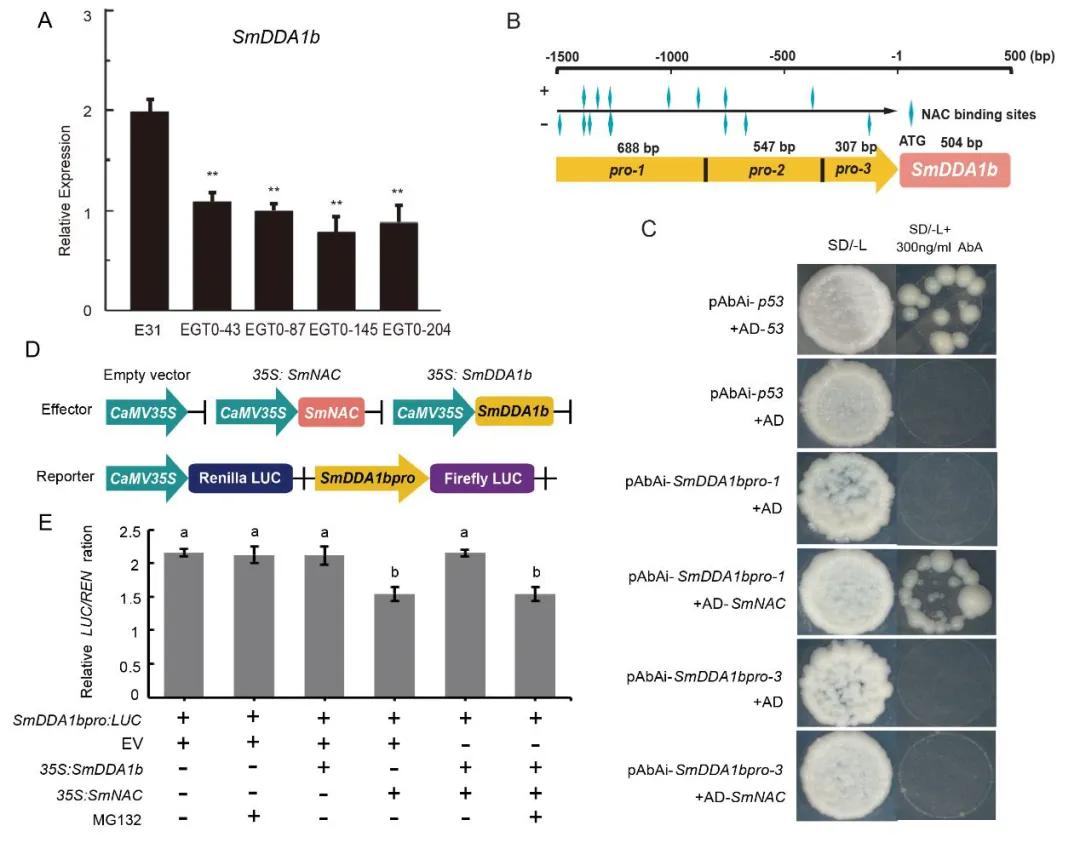 Figure 16 depicts the interaction between SmNAC and the promoter of SmDDA1b, along with the suppression of SmDDA1b expression (Yan et al., 2023). Panel A illustrates the expression levels of SmDDA1b in SmNAC-overexpressing plants, denoted as E31 for wild type, and EGT0-43, EGT0-87, EGT0-145, and EGT0-204 for T0 generation overexpressing plants. Panel B presents a schematic diagram of the truncated promoter. Panels C and D-E respectively represent the yeast one-hybrid (Y1H) assay and Dual-LUC assay, including the construction of vectors and the determination of enzyme activity.
Figure 16 depicts the interaction between SmNAC and the promoter of SmDDA1b, along with the suppression of SmDDA1b expression (Yan et al., 2023). Panel A illustrates the expression levels of SmDDA1b in SmNAC-overexpressing plants, denoted as E31 for wild type, and EGT0-43, EGT0-87, EGT0-145, and EGT0-204 for T0 generation overexpressing plants. Panel B presents a schematic diagram of the truncated promoter. Panels C and D-E respectively represent the yeast one-hybrid (Y1H) assay and Dual-LUC assay, including the construction of vectors and the determination of enzyme activity.
Based on the aforementioned experimental results, the authors constructed a model diagram illustrating the regulation of eggplant resistance to bacterial wilt by SmDDA1b (Figure 17). In resistant eggplants, during bacterial wilt pathogen stress, the expression of SmDDA1b protein is induced. SmNAC is recognized by SmDDA1b and subsequently degraded by the ubiquitin/26S proteasome system (UPS) mediated by SmDDA1b. This alleviates the inhibition of ICS1 expression, a gene involved in salicylic acid (SA) biosynthesis, thereby activating the SA signaling pathway and triggering the immune response in eggplants, ultimately enhancing their resistance to bacterial wilt. Conversely, in susceptible eggplants, the expression of SmDDA1b is suppressed during bacterial wilt pathogen stress. Consequently, SmNAC cannot be recognized and degraded by the UPS, and SmNAC protein, in turn, inhibits the expression of SmDDA1b. This enhances the inhibitory effect of SmNAC on SmICS1, leading to suppression of the SA signaling pathway, resulting in decreased resistance of eggplants to bacterial wilt.
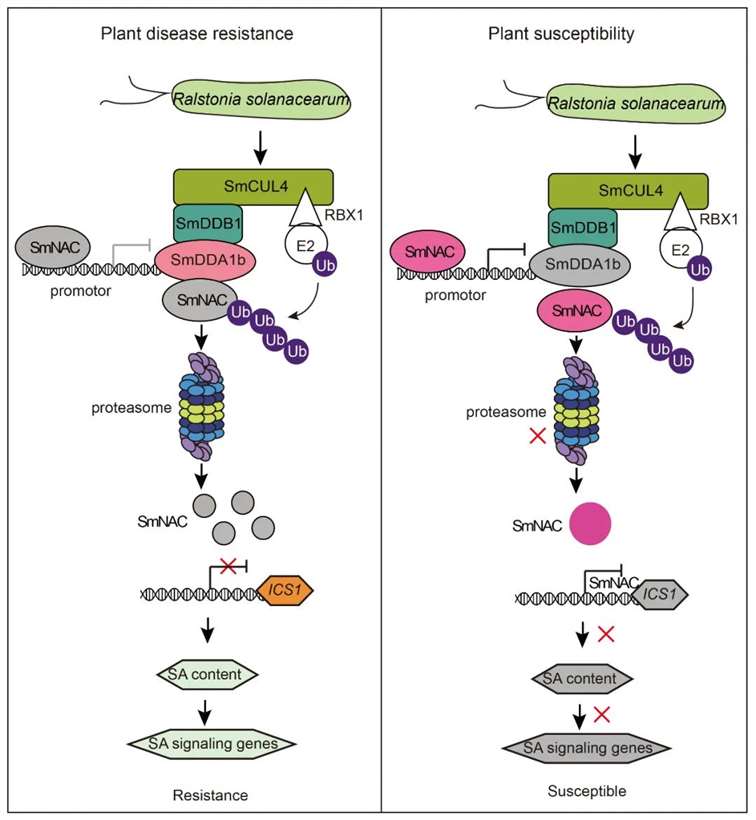 Figure 17 Model diagram of SmDDA1b regulating plant resistance to bacterial wilt disease (Yan et al., 2023).
Figure 17 Model diagram of SmDDA1b regulating plant resistance to bacterial wilt disease (Yan et al., 2023).
Participate in Plant Growth and Development
The plant hormone abscisic acid (ABA) plays a crucial role in regulating fruit maturation, with E3 ubiquitin ligases participating in plant growth and development by modulating ABA biosynthesis, transport, and signaling pathways.
A study titled "Grapevine U-Box E3 Ubiquitin Ligase VlPUB38 Negatively Regulates Fruit Ripening by Facilitating Abscisic-Aldehyde Oxidase Degradation" was published in the Plant and Cell Physiology journal. In this research, the authors demonstrated that the E3 ubiquitin ligase VlPUB38 mediates the degradation of abscisic-aldehyde oxidase (VvAAO) via the 26S proteasome, thereby regulating ABA synthesis and participating in the modulation of grapevine fruit ripening.
Initially, the authors analyzed the morphological and physiological parameters of grapevine fruit development across five stages. They observed a continuous increase in fruit diameter, with fruit hardness reaching its peak at the EL-34 stage before declining. Total soluble solids and anthocyanin content showed an increase across all five stages. Through qRT-PCR experiments, the authors found that the expression of VlPUB38 was significantly higher in fruit and inflorescence compared to other tissue types, with its expression decreasing at the EL-35 stage (the onset of fruit ripening) (Figure 18). These findings indicate the involvement of VlPUB38 in grapevine fruit development.
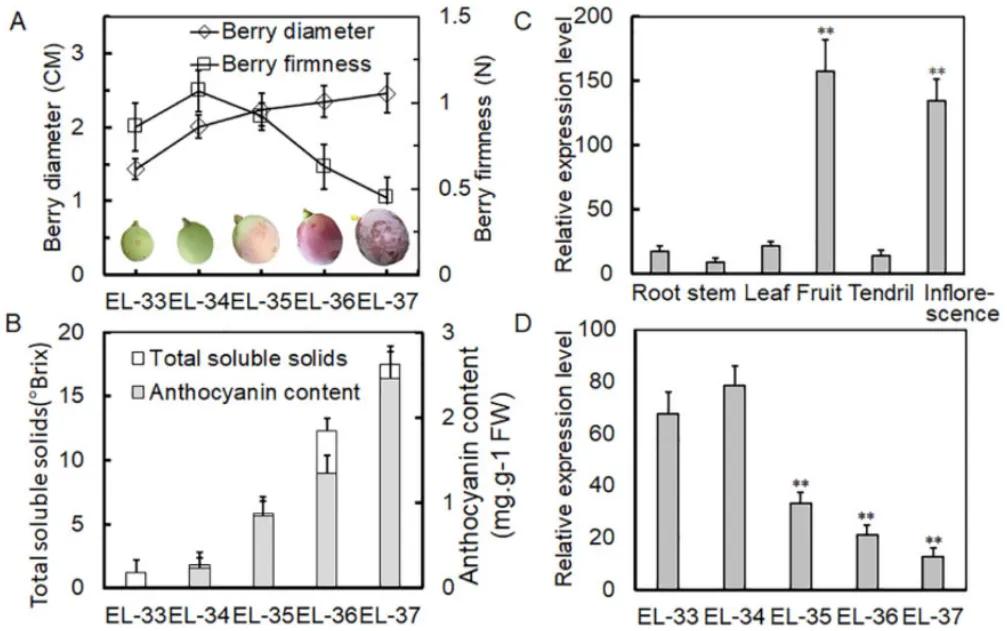 Figure 18 depicts the expression of VlPUB38 during grapevine fruit development (Yu et al., 2020). Panel (A) shows measurements of berry diameter and hardness, while panel (B) illustrates the determination of total soluble solids and anthocyanin content in the berries. Panel (C) presents the qRT-PCR expression analysis of VlPUB38 in various organs of grapevine plants, and panel (D) displays the qRT-PCR expression analysis of VlPUB38 at different developmental stages of the berries.
Figure 18 depicts the expression of VlPUB38 during grapevine fruit development (Yu et al., 2020). Panel (A) shows measurements of berry diameter and hardness, while panel (B) illustrates the determination of total soluble solids and anthocyanin content in the berries. Panel (C) presents the qRT-PCR expression analysis of VlPUB38 in various organs of grapevine plants, and panel (D) displays the qRT-PCR expression analysis of VlPUB38 at different developmental stages of the berries.
Plant U-box proteins exhibit E3 ubiquitin ligase activity. Through domain analysis, it was found that VlPUB38 contains a U-box domain. Therefore, the authors hypothesized that VlPUB38 functions as an E3 ubiquitin ligase. This hypothesis was confirmed by in vitro ubiquitin ligase activity assays, as depicted in Figure 19.
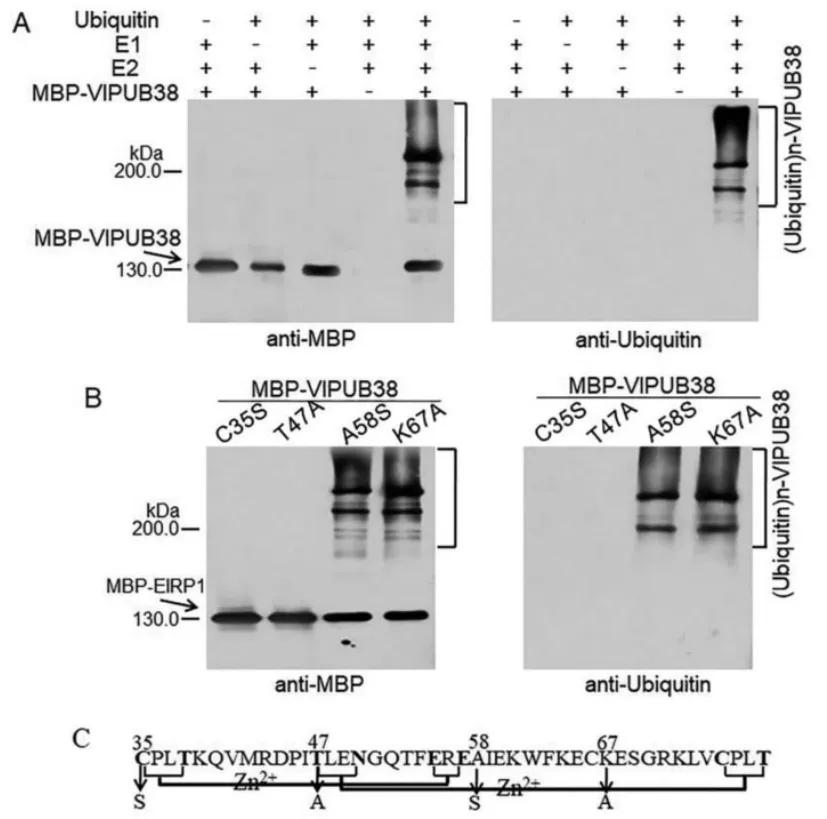 Figure 19 illustrates the E3 ubiquitin ligase activity of VlPUB38 (Yu et al., 2020). Panel A depicts the assay for the E3 ubiquitin ligase activity of VlPUB38, while Panel B shows the assay for the E3 ubiquitin ligase activity of four VlPUB38 mutants. Panel C presents a schematic diagram of the conserved domain and mutation sites of VlPUB38.
Figure 19 illustrates the E3 ubiquitin ligase activity of VlPUB38 (Yu et al., 2020). Panel A depicts the assay for the E3 ubiquitin ligase activity of VlPUB38, while Panel B shows the assay for the E3 ubiquitin ligase activity of four VlPUB38 mutants. Panel C presents a schematic diagram of the conserved domain and mutation sites of VlPUB38.
The transient overexpression vector of VlPUB38 was constructed by the authors, revealing that the overexpression of VlPUB38 inhibited the ripening of strawberry fruits. Simultaneously, upon ABA treatment of the strawberry fruits overexpressing VlPUB38, an increase in the content of soluble substances and anthocyanins was observed. Additionally, the expression of the key gene NCED1 involved in ABA synthesis was upregulated, while the expression of the key gene ABI1 in the ABA signaling pathway was downregulated (Figure 20). These findings collectively indicate that VlPUB38 suppresses the ripening of strawberry fruits and is involved in regulating ABA synthesis and signaling transduction processes.
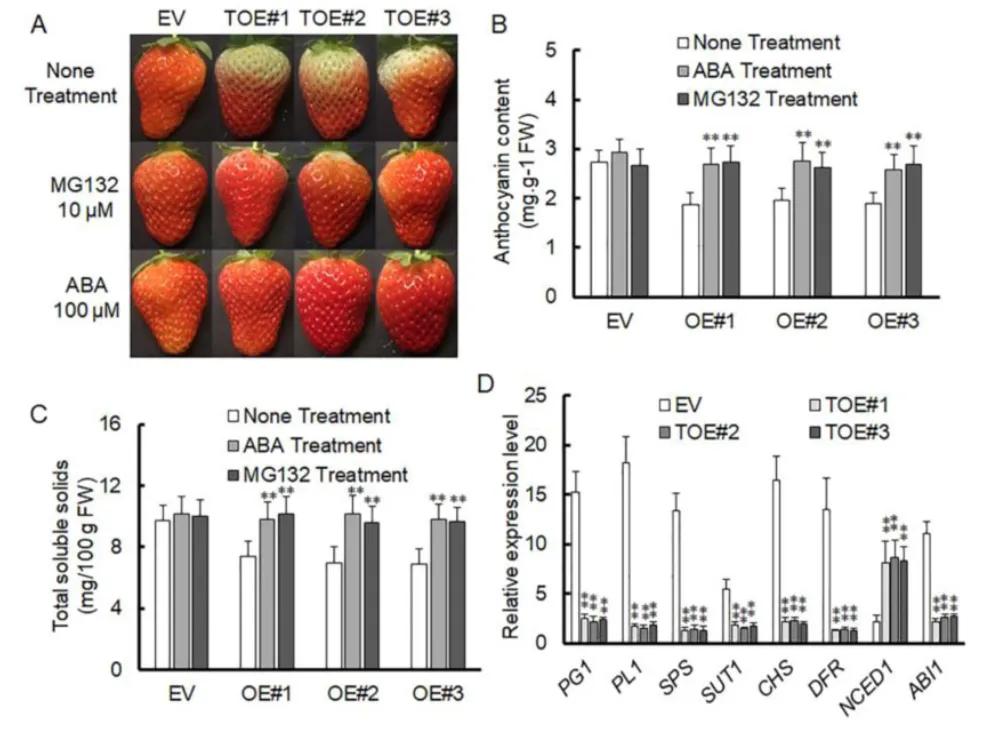 Figure 20 depicts the inhibition of strawberry fruit ripening by overexpressing VlPUB38 (Yu et al., 2020). Panel A illustrates the growth phenotypes of strawberries overexpressing VlPUB38 compared to wild-type strawberries. Panels B and C present the determination of anthocyanin and total soluble solids content in strawberries overexpressing VlPUB38. Panel D shows the qRT-PCR analysis of gene expression related to ripening in strawberries overexpressing VlPUB38.
Figure 20 depicts the inhibition of strawberry fruit ripening by overexpressing VlPUB38 (Yu et al., 2020). Panel A illustrates the growth phenotypes of strawberries overexpressing VlPUB38 compared to wild-type strawberries. Panels B and C present the determination of anthocyanin and total soluble solids content in strawberries overexpressing VlPUB38. Panel D shows the qRT-PCR analysis of gene expression related to ripening in strawberries overexpressing VlPUB38.
To further validate the function of VlPUB38, protein-protein interaction was assessed through yeast two-hybrid screening, combined with Bimolecular Fluorescence Complementation (BiFC) and Glutathione S-transferase (GST) Pull-down assays, confirming the interaction with the protein VIAAO (Figure 21). Basic bioinformatics analysis revealed VIAAO as a homologous protein of ABA-aldehyde oxidase in Arabidopsis thaliana. Subsequent experiments employing cell-free degradation assays and in vitro ubiquitination assays demonstrated that VlPUB38 mediates the ubiquitination degradation of VIAAO through the 26S proteasome (Figure 22).
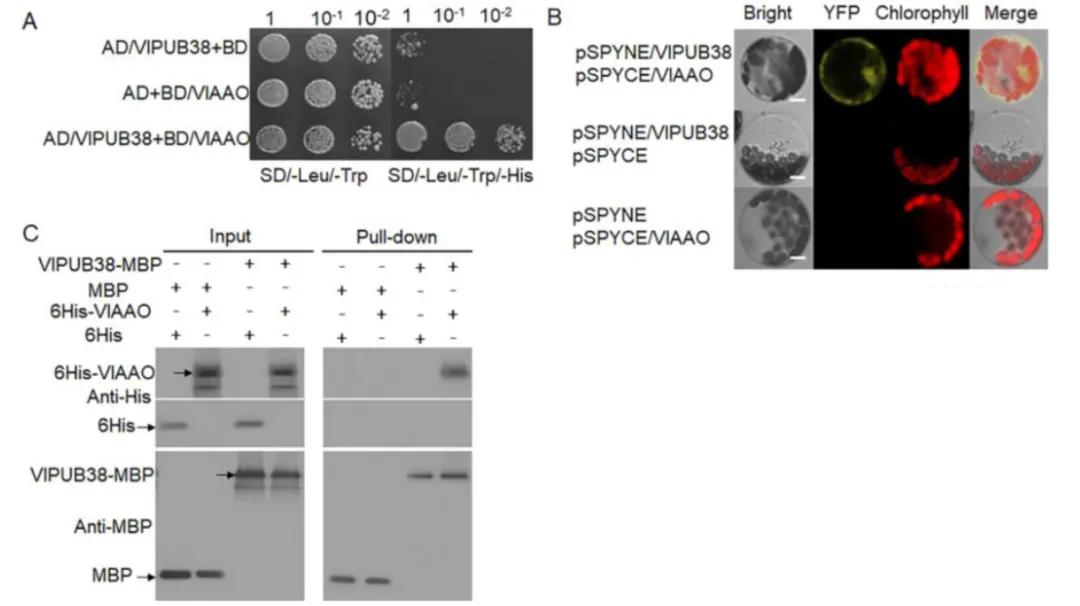 Figure 21 depicts the interaction between VlPUB38 and VlAAO (Yu et al., 2020). The interaction was validated through (A) yeast two-hybrid (Y2H) assay, (B) Bimolecular Fluorescence Complementation (BiFC) assay, and (C) Glutathione S-transferase (GST) Pull-down assay.
Figure 21 depicts the interaction between VlPUB38 and VlAAO (Yu et al., 2020). The interaction was validated through (A) yeast two-hybrid (Y2H) assay, (B) Bimolecular Fluorescence Complementation (BiFC) assay, and (C) Glutathione S-transferase (GST) Pull-down assay.
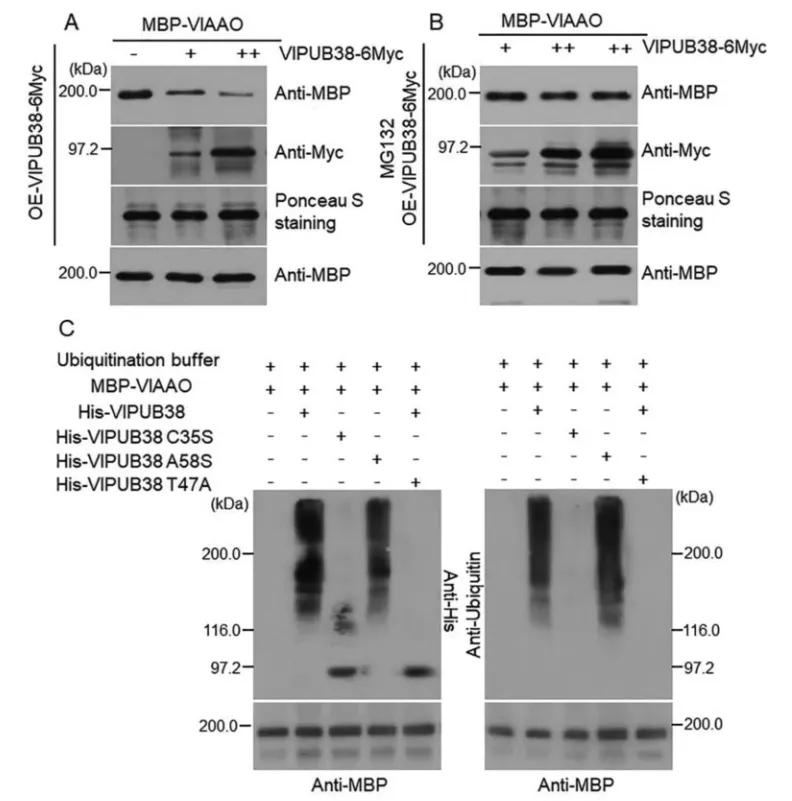 Figure 22 illustrates the ubiquitination and degradation of VlAAO mediated by VlPUB38 through the 26S proteasome (Yu et al., 2020). The (A, B) cell-free degradation experiments demonstrate that overexpression of VlPUB38 leads to reduced levels of VlAAO expression, with MG132 serving as a 26S proteasome inhibitor. (C) VlPUB38 facilitates the ubiquitination of VlAAO.
Figure 22 illustrates the ubiquitination and degradation of VlAAO mediated by VlPUB38 through the 26S proteasome (Yu et al., 2020). The (A, B) cell-free degradation experiments demonstrate that overexpression of VlPUB38 leads to reduced levels of VlAAO expression, with MG132 serving as a 26S proteasome inhibitor. (C) VlPUB38 facilitates the ubiquitination of VlAAO.
Based on the aforementioned experimental findings, a model diagram depicting the regulatory role of VlPUB38 in grape fruit development was constructed by the authors (Figure 23). VlPUB38 functions as a negative regulator of grape fruit ripening, with its encoded protein capable of recognizing and ubiquitinating VlAAO. Subsequently, VlAAO undergoes hydrolysis via the 26S proteasome system, thereby modulating ABA levels and ultimately regulating fruit development.
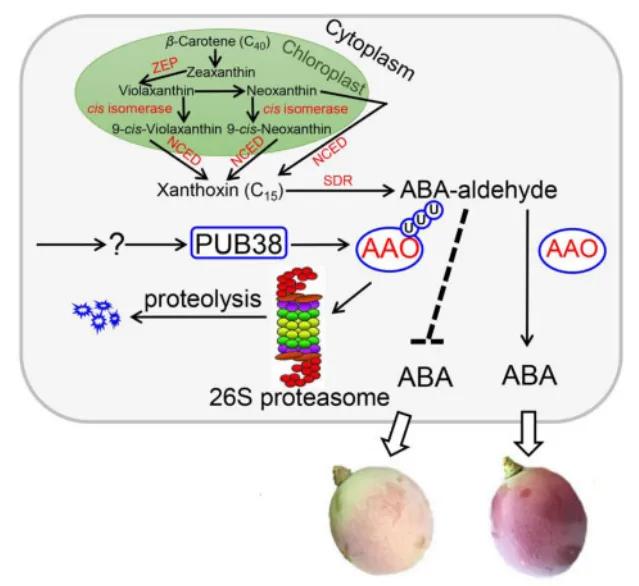 Figure 23 Model diagram of VlPUB38 regulating grape fruit development (Yu et al., 2020).
Figure 23 Model diagram of VlPUB38 regulating grape fruit development (Yu et al., 2020).
References
- Chen, S., Xu, K., Kong, D., et al. (2022). Ubiquitin ligase OsRINGzf1 regulates drought resistance by controlling the turnover of OsPIP2; 1. Plant Biotechnology Journal, 20(9), 1743-1755.
- Dittmar, G., Winklhofer, K. F. (2020). Linear ubiquitin chains: cellular functions and strategies for detection and quantification. Frontiers in Chemistry, 7, 915.
- Fennell, L. M., Rahighi, S., Ikeda, F. (2018). Linear ubiquitin chain‐binding domains. The FEBS Journal, 285(15), 2746-2761.
- Franklin, T. G., Pruneda, J. N. (2019). A high-throughput assay for monitoring ubiquitination in real time. Frontiers in Chemistry, 7, 816.
- Irigoyen, M. L., Iniesto, E., Rodriguez, L., et al. (2014). Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. The Plant Cell, 26(2), 712-728.
- Ma, X., Zhang, C., Kim, D. Y., et al. (2021). Ubiquitylome analysis reveals a central role for the ubiquitin-proteasome system in plant innate immunity. Plant physiology, 185(4), 1943-1965.
- McClellan, A. J., Laugesen, S. H., Ellgaard, L. (2019). Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biology, 9(9), 190147.
- Nandi, D., Tahiliani, P., Kumar, A., et al. (2006). The ubiquitin-proteasome system. Journal of biosciences, 31(1), 137-155.
- Na, C., Shuanghua, W., Jinglong, F., et al. (2016). Overexpression of the Eggplant (Solanum melongena) NAC Family Transcription Factor SmNAC Suppresses Resistance to Bacterial Wilt. Scientific Reports, 6, 31568.
- Pang, P. X., Shi, L., Wang, X. J., et al. (2019). Cloning and expression analysis of the StCUL1 gene in potato. Journal of Plant Biochemistry and Biotechnology, 28, 460-469.
- Pickart, C. M., Fushman, D. (2004). Polyubiquitin chains: polymeric protein signals. Current opinion in chemical biology, 8(6), 610-616.
- Ruan, W., Guo, M., Wang, X., et al. (2019). Two RING-finger ubiquitin E3 ligases regulate the degradation of SPX4, an internal phosphate sensor, for phosphate homeostasis and signaling in rice. Molecular plant, 12(8), 1060-1074.
- Saracco, S. A., Hansson, M., Scalf, M., et al. (2009). Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. The Plant Journal, 59(2), 344-358.
- Sowa, M. E., Bennett, E. J., Gygi, S. P., et al. (2009). Defining the human deubiquitinating enzyme interaction landscape. Cell, 138(2), 389-403.
- Stone, S. L. (2019). Role of the ubiquitin proteasome system in plant response to abiotic stress. International review of cell and molecular biology, 343, 65-110.
- Swatek, K. N., Komander, D. (2016). Ubiquitin modifications. Cell research, 26(4), 399-422.
- Xiao, W., Liu, Z., Luo, W., et al. (2019). Specific and unbiased detection of polyubiquitination via a sensitive non-antibody approach. Analytical Chemistry, 92(1).
- Xin, Xie, Hou xiang, et al. (2015). Comprehensive Profiling of the Rice Ubiquitome Reveals the Significance of Lysine Ubiquitination in Young Leaves. Journal of Proteome Research, 14(5), 2017-2025.
- Yan, S., Wang, Y., Yu, B., et al. (2023). A putative E3 ubiquitin ligase substrate receptor degrades transcription factor SmNAC to enhance bacterial wilt resistance in Eggplant. Horticulture Research, uhad246.
- Yu, Y., Meng, X., Guo, D., et al. (2020). Grapevine U-box E3 ubiquitin ligase VlPUB38 negatively regulates fruit ripening by facilitating abscisic-aldehyde oxidase degradation. Plant and Cell Physiology, 61(12), 2043-2054.
- Zhang, Y., Zeng, L. (2020). Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity. Plant communications, 1(4).
- Zhu, L., Cheng, H., Peng, G., et al. (2020). Ubiquitinome Profiling Reveals the Landscape of Ubiquitination Regulation in Rice Young Panicles. Genomics Proteomics & Bioinformatics.
Our products and services are for research use only.


 Figure 1 depicts a heterotypic ubiquitin chain containing the M1 linkage (Gunnar Dittmar and Konstanze F. Winklhofer, 2020).
Figure 1 depicts a heterotypic ubiquitin chain containing the M1 linkage (Gunnar Dittmar and Konstanze F. Winklhofer, 2020). Figure 2. Ubiquitination Process (Stone, Sophia L., 2019) illustrates the ubiquitination process. The following abbreviations are used in the figure: E1 denotes the ubiquitin-activating enzyme; E2 represents the ubiquitin-conjugating enzyme; E3 signifies the ubiquitin-ligase enzyme; Target indicates the substrate protein; Ubiquitin refers to the ubiquitin molecule; and DUBs represent deubiquitinating enzymes.
Figure 2. Ubiquitination Process (Stone, Sophia L., 2019) illustrates the ubiquitination process. The following abbreviations are used in the figure: E1 denotes the ubiquitin-activating enzyme; E2 represents the ubiquitin-conjugating enzyme; E3 signifies the ubiquitin-ligase enzyme; Target indicates the substrate protein; Ubiquitin refers to the ubiquitin molecule; and DUBs represent deubiquitinating enzymes. Figure 3: Physiological Functions Associated with Different Types of Ubiquitin Chains (Kirby N Swatek and David Komander, 2016)
Figure 3: Physiological Functions Associated with Different Types of Ubiquitin Chains (Kirby N Swatek and David Komander, 2016) Figure 4 Formation of 26S proteasome, this process requires the consumption of ATP (Nandi et al., 2006).
Figure 4 Formation of 26S proteasome, this process requires the consumption of ATP (Nandi et al., 2006). Figure 5: Characteristics of Identified Lysine-Ubiquitinated Peptides in Rice Young Panicles (Zhu et al., 2020)
Figure 5: Characteristics of Identified Lysine-Ubiquitinated Peptides in Rice Young Panicles (Zhu et al., 2020) Figure 6: Ubiquitin Ligase Activity of OsRINGzf1 (Chen et al., 2022)
Figure 6: Ubiquitin Ligase Activity of OsRINGzf1 (Chen et al., 2022) Figure 7: Ubiquitination of OsPIP2;1 Mediated by OsRINGzf1 (Chen et al., 2022)
Figure 7: Ubiquitination of OsPIP2;1 Mediated by OsRINGzf1 (Chen et al., 2022) Figure 8: Overview of UbiReal Experimental Principle (Franklin and Pruneda, 2019)
Figure 8: Overview of UbiReal Experimental Principle (Franklin and Pruneda, 2019) Figure 9 A new technology for detecting protein ubiquitination (TUF-WB) formed by ThUBD combined with Far-Western blot (Xiao et al., 2019).
Figure 9 A new technology for detecting protein ubiquitination (TUF-WB) formed by ThUBD combined with Far-Western blot (Xiao et al., 2019). Figure 10: Specific Recognition of Ubiquitinated Proteins by TUF-WB (Xiao et al., 2019).
Figure 10: Specific Recognition of Ubiquitinated Proteins by TUF-WB (Xiao et al., 2019). Figure 11: Affinity Purification of Ubiquitinated Proteins Expressing 6HIS-UBQ in Transgenic Arabidopsis thaliana (Ma et al., 2021).
Figure 11: Affinity Purification of Ubiquitinated Proteins Expressing 6HIS-UBQ in Transgenic Arabidopsis thaliana (Ma et al., 2021). Figure 12: Gene Ontology (GO) Analysis of Ubiquitinated Proteins in flg22-Treated and Untreated Samples (Ma et al., 2021).
Figure 12: Gene Ontology (GO) Analysis of Ubiquitinated Proteins in flg22-Treated and Untreated Samples (Ma et al., 2021). Figure 13: Interaction between SmDDA1b and SmNAC and Subcellular Localization of SmDDA1b (Yan et al., 2023).
Figure 13: Interaction between SmDDA1b and SmNAC and Subcellular Localization of SmDDA1b (Yan et al., 2023). Figure 14: Positive Regulation of Salicylic Acid (SA) Biosynthesis and Participation in SA Signaling Pathways by SmDDA1b (Yan et al., 2023).
Figure 14: Positive Regulation of Salicylic Acid (SA) Biosynthesis and Participation in SA Signaling Pathways by SmDDA1b (Yan et al., 2023). Figure 15: Interaction between SmDDB1, SmCUL4, and SmDDA1b, and SmDDA1b-Mediated Degradation of SmNAC (Yan et al., 2023).
Figure 15: Interaction between SmDDB1, SmCUL4, and SmDDA1b, and SmDDA1b-Mediated Degradation of SmNAC (Yan et al., 2023). Figure 16 depicts the interaction between SmNAC and the promoter of SmDDA1b, along with the suppression of SmDDA1b expression (Yan et al., 2023). Panel A illustrates the expression levels of SmDDA1b in SmNAC-overexpressing plants, denoted as E31 for wild type, and EGT0-43, EGT0-87, EGT0-145, and EGT0-204 for T0 generation overexpressing plants. Panel B presents a schematic diagram of the truncated promoter. Panels C and D-E respectively represent the yeast one-hybrid (Y1H) assay and Dual-LUC assay, including the construction of vectors and the determination of enzyme activity.
Figure 16 depicts the interaction between SmNAC and the promoter of SmDDA1b, along with the suppression of SmDDA1b expression (Yan et al., 2023). Panel A illustrates the expression levels of SmDDA1b in SmNAC-overexpressing plants, denoted as E31 for wild type, and EGT0-43, EGT0-87, EGT0-145, and EGT0-204 for T0 generation overexpressing plants. Panel B presents a schematic diagram of the truncated promoter. Panels C and D-E respectively represent the yeast one-hybrid (Y1H) assay and Dual-LUC assay, including the construction of vectors and the determination of enzyme activity. Figure 17 Model diagram of SmDDA1b regulating plant resistance to bacterial wilt disease (Yan et al., 2023).
Figure 17 Model diagram of SmDDA1b regulating plant resistance to bacterial wilt disease (Yan et al., 2023). Figure 18 depicts the expression of VlPUB38 during grapevine fruit development (Yu et al., 2020). Panel (A) shows measurements of berry diameter and hardness, while panel (B) illustrates the determination of total soluble solids and anthocyanin content in the berries. Panel (C) presents the qRT-PCR expression analysis of VlPUB38 in various organs of grapevine plants, and panel (D) displays the qRT-PCR expression analysis of VlPUB38 at different developmental stages of the berries.
Figure 18 depicts the expression of VlPUB38 during grapevine fruit development (Yu et al., 2020). Panel (A) shows measurements of berry diameter and hardness, while panel (B) illustrates the determination of total soluble solids and anthocyanin content in the berries. Panel (C) presents the qRT-PCR expression analysis of VlPUB38 in various organs of grapevine plants, and panel (D) displays the qRT-PCR expression analysis of VlPUB38 at different developmental stages of the berries. Figure 19 illustrates the E3 ubiquitin ligase activity of VlPUB38 (Yu et al., 2020). Panel A depicts the assay for the E3 ubiquitin ligase activity of VlPUB38, while Panel B shows the assay for the E3 ubiquitin ligase activity of four VlPUB38 mutants. Panel C presents a schematic diagram of the conserved domain and mutation sites of VlPUB38.
Figure 19 illustrates the E3 ubiquitin ligase activity of VlPUB38 (Yu et al., 2020). Panel A depicts the assay for the E3 ubiquitin ligase activity of VlPUB38, while Panel B shows the assay for the E3 ubiquitin ligase activity of four VlPUB38 mutants. Panel C presents a schematic diagram of the conserved domain and mutation sites of VlPUB38. Figure 20 depicts the inhibition of strawberry fruit ripening by overexpressing VlPUB38 (Yu et al., 2020). Panel A illustrates the growth phenotypes of strawberries overexpressing VlPUB38 compared to wild-type strawberries. Panels B and C present the determination of anthocyanin and total soluble solids content in strawberries overexpressing VlPUB38. Panel D shows the qRT-PCR analysis of gene expression related to ripening in strawberries overexpressing VlPUB38.
Figure 20 depicts the inhibition of strawberry fruit ripening by overexpressing VlPUB38 (Yu et al., 2020). Panel A illustrates the growth phenotypes of strawberries overexpressing VlPUB38 compared to wild-type strawberries. Panels B and C present the determination of anthocyanin and total soluble solids content in strawberries overexpressing VlPUB38. Panel D shows the qRT-PCR analysis of gene expression related to ripening in strawberries overexpressing VlPUB38. Figure 21 depicts the interaction between VlPUB38 and VlAAO (Yu et al., 2020). The interaction was validated through (A) yeast two-hybrid (Y2H) assay, (B) Bimolecular Fluorescence Complementation (BiFC) assay, and (C) Glutathione S-transferase (GST) Pull-down assay.
Figure 21 depicts the interaction between VlPUB38 and VlAAO (Yu et al., 2020). The interaction was validated through (A) yeast two-hybrid (Y2H) assay, (B) Bimolecular Fluorescence Complementation (BiFC) assay, and (C) Glutathione S-transferase (GST) Pull-down assay. Figure 22 illustrates the ubiquitination and degradation of VlAAO mediated by VlPUB38 through the 26S proteasome (Yu et al., 2020). The (A, B) cell-free degradation experiments demonstrate that overexpression of VlPUB38 leads to reduced levels of VlAAO expression, with MG132 serving as a 26S proteasome inhibitor. (C) VlPUB38 facilitates the ubiquitination of VlAAO.
Figure 22 illustrates the ubiquitination and degradation of VlAAO mediated by VlPUB38 through the 26S proteasome (Yu et al., 2020). The (A, B) cell-free degradation experiments demonstrate that overexpression of VlPUB38 leads to reduced levels of VlAAO expression, with MG132 serving as a 26S proteasome inhibitor. (C) VlPUB38 facilitates the ubiquitination of VlAAO. Figure 23 Model diagram of VlPUB38 regulating grape fruit development (Yu et al., 2020).
Figure 23 Model diagram of VlPUB38 regulating grape fruit development (Yu et al., 2020).