Top-Down Mass Spectrometry Service
Online InquiryTop-down mass spectrometry is an analytical strategy for identifying proteins, characterizing post-translational modifications, and even quantifying proteins. This method allows analysis of intact proteins without the need to digest peptides.
With its advanced mass spectrometry platform and rich experience in proteomics analysis, Creative Proteomics can provide you with top-down mass spectrometry services for the study of organelle and subcellular structure.
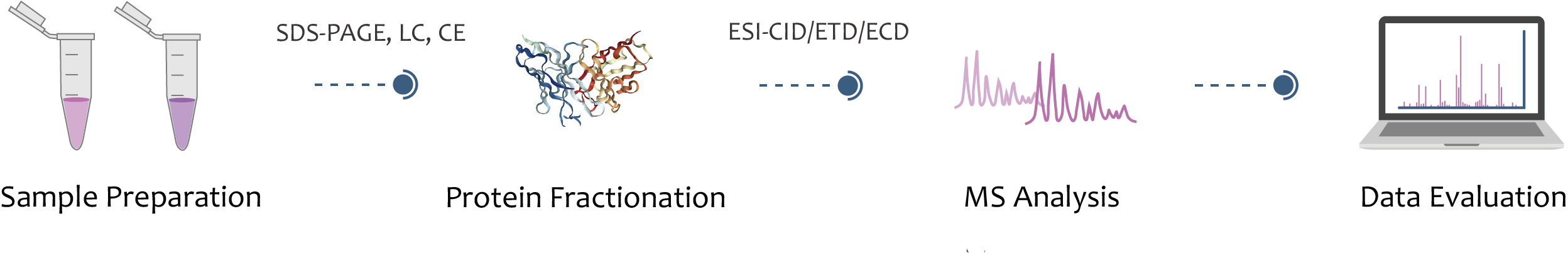
Top-Down Mass Spectrometry Principle and Workflow
Top-down mass spectrometry mainly includes complete protein separation technology, mass spectrometry technology and bioinformatics analysis. The separation of intact proteins is to reduce the complexity of sample injection for mass spectrometry, and to improve the "purity" and signal-to-noise ratio of mass spectrometry data. Creative Proteomics offers subcellular structure isolation and protein extraction and purification to help you obtain complete proteins for subsequent top-down mass spectrometric analysis.
Complete proteins are ionized by electrospray ionization (ESI) or laser matrix desorption ionization (MALDI). The generated ions are fragmented by collision induced dissociation (CID), electron capture dissociation (ECD), or electron transfer dissociation (ETD), and analyzed in tandem mass spectrometry to obtain protein information.
Technology Platform
Q Exactive Hybrid Quadrupole-Orbitrap and Agilent 6540 Q-TOF high performance mass spectrometers are available.
Advantages of Top-Down Mass Spectrometry:
- Does not require the laborious chemical or enzymatic digestion.
- 100% sequence coverage and full characterization of proteoforms.
- Quick characterization of N-terminus and C-terminus
- PTM combinations on large segments can be detected and positional isomers with the same molecular weight can be distinguished. Protein-level variation and membrane protein analysis are feasible.
Bioinformatics Analysis
| Qualitative Analysis of Protein | Data Quality Control | Precursor Ion Mass Tolerance Distribution |
| Peptide Length Distribution | ||
| Unique Peptide Number Distribution | ||
| Protein Coverage Distribution | ||
| Protein Function Annotation | GO Annotation | |
| COG Annotation | ||
| KEGG Annotation | ||
| Domain Annotation | ||
| Quantitative Analysis of Protein | Protein Differential Analysis | Ratio Analysis |
| Volcano Map Analysis | ||
| Expression Cluster Analysis | ||
| Functional Enrichment Analysis of Differential Proteins | GO Enrichment Analysis | |
| KEGG Enrichment Analysis | ||
| Domain Enrichment Analysis | ||
| Protein Interaction Network Analysis |
Delivery
Experimental procedures, liquid chromatography and mass spectrometer parameters, MS raw data files, bioinformatics analysis charts. Reports are available in Excel or PDF format.
Want to Know about Other Protein Identification and/or Quantification Service?
References
- Kar U K, Simonian M, Whitelegge J P. Integral membrane proteins: bottom-up, top-down and structural proteomics. Expert review of proteomics, 2017, 14(8): 715-723.
- Van De Waterbeemd M, Tamara S, Fort K L, et al. Dissecting ribosomal particles throughout the kingdoms of life using advanced hybrid mass spectrometry methods. Nature communications, 2018, 9(1): 2493.
- Siuti N, Kelleher N L. Decoding protein modifications using top-down mass spectrometry. Nature methods, 2007, 4(10): 817.
- Di Pierro A, Bondi H, Monti C, et al. Experimental setup for the identification of mitochondrial protease substrates by shotgun and top-down proteomics. EuPA open proteomics, 2016, 11: 1-3.
* For Research Use Only. Not for use in diagnostic procedures.