Exosomes Lipidomics Service
Online Inquiry- Service Details
- Case Study
Lipids are an important part of exosomes. A number of studies have shown that lipids play an important role in exosomal biology. Compared with donor cells, exosomes are more enriched in sphingolipids, cholesterol and phosphatidylserine. The lipid composition of exosomes is similar to lipid rafts, and exosomes have higher lipid content than other extracellular vesicles and higher stability to detergents. Improving the understanding of lipids will help us better understand the biological characteristics of exosomes. Exosomal lipidomics is an urgent need now.
Creative Proteomics aims to provide high-quality exosomes lipidomics services for customers in different fields. Our scientists help speed up your project progress based on professional knowledge and high-sensitivity equipment. All you need is to send us your samples or purified exosomes. We will deliver you a report containing identification, mass/charge ratio and metabolite difference analysis.
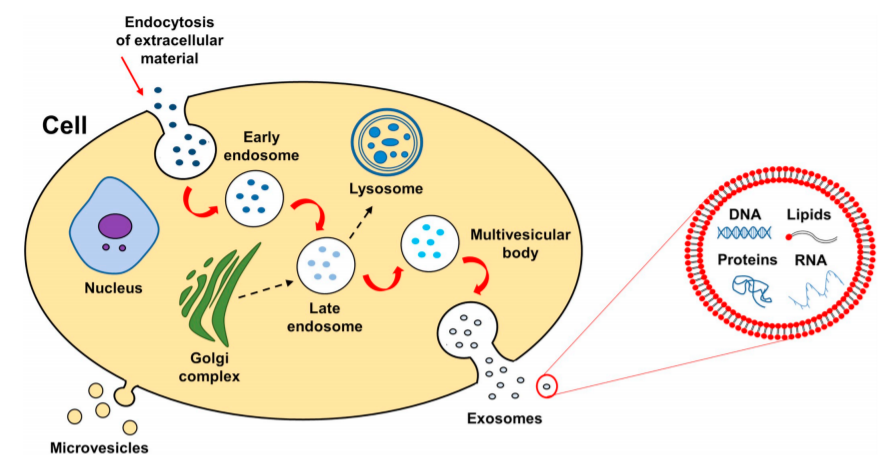
Exosomes Lipidomics Service in Creative Proteomics
Targeted lipidomics: targeted detection and quantification of a specific lipid or a certain class of lipids.
Untargeted lipidomics: Based on GC-MS and LC-MS, nonbiased detection of all lipid molecules in as many biological samples as possible, such as cells, tissues, organs, or body fluids. The experimental group and the control group are compared and analyzed. Differential lipid molecules are screened through statistical analysis, and the relative relationship between lipid metabolism and physiological and pathological changes is found, so as to reveal the functional mechanism of lipids in various life activities.
1. Analysis and identification of lipids and their metabolites
2. Study on lipid metabolic pathway and network
3. Regulation of lipid function and metabolism
Detected Lipids for Exosome Lipidomics
| Lipid Class | Typical Lipid Species |
|---|---|
| Phospholipids | Phosphatidylcholine (PC), Phosphatidylethanolamine (PE), Phosphatidylinositol (PI) |
| Glycolipids | Diglycerides (DG), Triglycerides (TG) |
| Sterols | Cholesterol, Sterol esters |
| Sphingolipids | Sphingomyelin, Ceramides, Sphingosine-1-phosphate |
| Fatty Acids | Free Fatty Acids, Glycerolipids |
| Ether Lipids | Ether-linked Phospholipids, Ether-linked Glycerolipids, Plasmalogens |
| Lysophospholipids | Lysophosphatidylcholine (LPC), Lysophosphatidylethanolamine (LPE) |
| Neutral Lipids | Monoacylglycerols (MAG), Diacylglycerols (DAG), Triacylglycerols (TAG) |
| Prenol Lipids | Coenzyme Q, Dolichols |
| Alkylglycerols | Ether-linked Diacylglycerols (eDAG), Ether-linked Triacylglycerols (eTAG) |
| Bioactive Lipids | Anandamide, 2-Arachidonoylglycerol (2-AG) |
| Ceramides | Ceramide (Cer), Ceramide-1-phosphate (C1P) |
| Lysosphingolipids | Lysosphingomyelin |
Advantage of exosomes lipidomics service
High accuracy of lipid molecular identification: compare the identification of the secondary mass spectrogram, the identification number is about 1000, and carry out the multi-level analysis of lipid categories, subtypes, species, and molecules.
Ultra-high resolution mass spectrometer platform: use QE series spectrometer for detection, the instrument has high resolution, high quality accuracy and good stability.
Standardized platform and strict quality control: full-process standard operating procedures (SOP) guidance, dual quality control of isotope internal standards and QC.
Rich project experience: high-throughput automated sample preparation system for metabolite extraction, real-time monitoring of instrument detection process.
Bioinformatics analysis
- Standard data analysis
- Lipid identification, quantification and composition analysis: lipid subclass and molecular identification and quantification, lipid subclass content ratio, lipid distribution range
- Lipid content difference analysis: total, Class level, Species level analysis
- Lipid chain length change analysis, lipid chain saturation change analysis
- Advanced data analysis
- Marker screening: integrated machine learning
- Correlation analysis with target parameters: OPLS regression analysis/WGCNA analysis
Creative Proteomics is based on a highly stable, reproducible and highly sensitive separation, characterization, identification and quantitative analysis system to provide you with a reliable, fast and cost-effective one-stop exosomes lipidomics analysis service. Thanks for your trust. If you have any questions or need other exosome analysis related services, please contact us.
Case: Differential Lipid Composition in Prostate Cancer Cells and Exosomes
Background
The study investigates the lipid composition of exosomes and the parent PC-3 cells, aiming to understand membrane structure, exosome stability, and potential implications for cell-to-cell communication. The focus is on elucidating the molecular details of lipidomes and exploring their significance in cancer diagnostics.
Samples
The study utilizes the epithelial human prostate cancer cell line PC-3 and the exosomes released by these cells. PC-3 cells are maintained in specific culture conditions, and both cells and exosomes undergo detailed experimental procedures for analysis.
Technical Methods
1. Materials: DMEM/F-12 medium, protein assay kit, protease inhibitor cocktail, etc.
2. Cell Culture: PC-3 cells maintained in a 1:1 mixture of Ham's F12 and DMEM.
3. Cell Collection: Cells trypsinized, washed, and stored at -80 °C.
4. Cell Lysates: Cells lysed in a buffer, and insoluble material removed by centrifugation.
5. Exosome Isolation: Exosomes isolated from serum-free culture medium using ultracentrifugation and characterized by Western blot and electron microscopy.
6. Protein Determination: Cells and exosomes lysed, and protein content determined.
7. Lipid Extraction: Lipids extracted from cells and exosomes using the Folch procedure.
8. Mass Spectrometry (MS) Analyses: Glycerophospholipids, glycerolipids, sterols, and sphingomyelin analyzed by shotgun analysis on a QTRAP 5500 mass spectrometer. Neutral glycosphingolipids analyzed by UHPLC. Ganglioside analysis performed using a modification of a published method.
9. Data Processing: MS data files processed using Lipid Profiler™ and MultiQuant™ software for lipid names and peak areas. Quality control samples used to monitor analysis quality.
10. Annotations of Lipid Species: Lipid species annotated, and concentrations presented as nmol/mg protein and mol% of total lipids.
Results
Analysis of Lipid Classes: Exosomes and PC-3 cells show remarkable differences in lipid composition. Exosomes are enriched in cholesterol and sphingomyelin, exhibiting a unique lipidome compared to parent cells.
Total Lipid Amounts: Exosomes contain an 8.4 times higher ratio of lipids/proteins than PC-3 cells, with most lipid classes elevated in exosomes.
Saturation Levels: Exosomes contain more saturated and less monounsaturated fatty acids than cells, and lipid classes in the outer and inner leaflets show distinct saturation patterns.
Analysis of Lipid Species: Specific lipid species are selectively enriched in exosomes, with significant differences observed in various classes such as sphingomyelin, ceramides, phosphatidylcholine, and phosphatidylserine.
Enrichment of Specific Lipids: Cholesterol, long-chain sphingolipids, and PS 18:0/18:1 are enriched to a similar extent in exosomes, suggesting potential interactions in the stability of membrane leaflets.
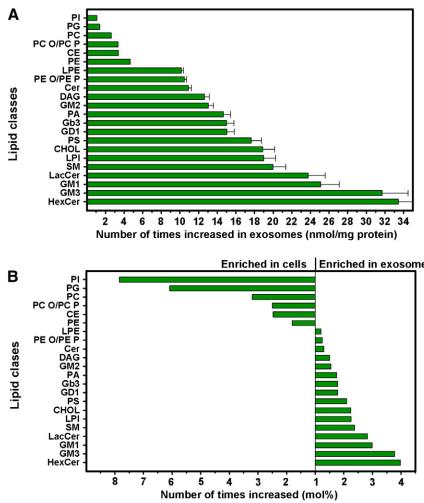 Lipid class enrichment in PC-3 cells and exosomes
Lipid class enrichment in PC-3 cells and exosomes
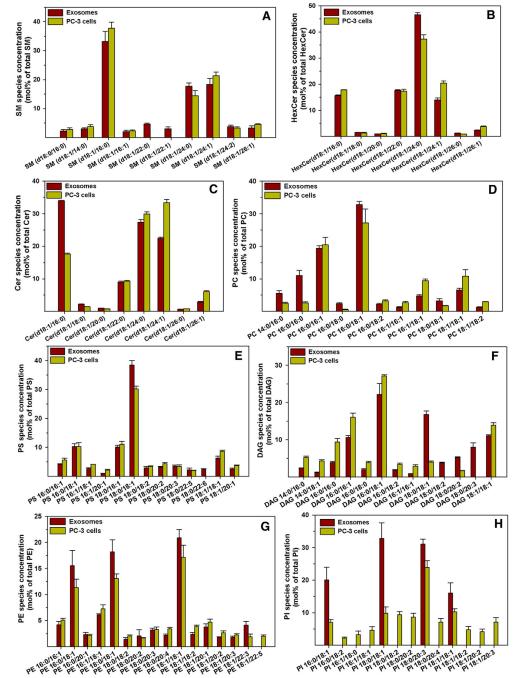 Lipid species composition of PC-3 cells and exosomes.
Lipid species composition of PC-3 cells and exosomes.
Reference
- Llorente, Alicia, et al. "Molecular lipidomics of exosomes released by PC-3 prostate cancer cells." Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids 1831.7 (2013): 1302-1309.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.