Protein Sequencing Service
Online Inquiry- Service Details
- Case Study
Protein molecules play a key role in the scientific research of biochemistry, kit development, diagnostic reagent development and drug development. The protein primary sequence determines the high-level structure of the protein and affects the function of the protein. Accurate and rapid analysis of protein sequences to verify the integrity and correct translation of protein samples is also helpful for the function research of unknown proteins.
Creative Proteomics aims to provide high-quality proteomics-related services to customers in various fields. Our scientists develop a protein sequencing platform based on professional knowledge and advanced equipment, which can provide accurate and reliable protein N/C terminal sequencing, protein de novo sequencing and mutation analysis, and protein full spectrum analysis.
Protein N/C terminal sequencing service
Determine the amino acid sequence at the N- and/or C-terminus of a protein.
- Edman degradation method: N-terminal sequence analysis of protein samples. The Edman degradation method cannot be used to analyze the protein sequence when the N-terminal is blocked or chemically modified.
- Mass spectrometry: The N-terminal sequence and C-terminal sequence of the protein can be determined simultaneously. The N-terminal sequencing technology based on mass spectrometry can realize the sequence determination of N-terminal blocked and PEGylated proteins, which is complementary to Edman sequencing. This analysis can be used to confirm whether the recombinant protein is fully expressed, to detect whether the recombinant protein expression process is broken, and whether the N-terminal and C-terminal sequences of the recombinant protein are modified.
- Sample requirement: 5-10ug/protein.
De novo protein sequencing and mutation analysis service
Full protein sequence coverage can be achieved. Suitable for sequence analysis of novel proteins that have not been previously reported, or proteins that have been purified from a novel species, as well as for sequence and mutation analysis of unknown proteins.
- Mass spectrometry: High-resolution Nano HPLC-Orbitrap Fusion Lumos Tribrid? Mass Spectrometer enables rapid and accurate determination of protein sequences.
- Sample requirement: Normal amount: 500ug/protein. Minimum amount: 200ug/protein.

Protein full spectrum analysis
It is mainly used for the comprehensive analysis of the protein species contained in unknown samples. Full spectrum protein analysis based on mass spectrometry technology can obtain as much protein information as possible, which can provide reference information for high-throughput quantitative and modification analysis of proteins. A systematic and comprehensive study of as many proteins as possible in a tissue or cell can identify up to 6,000 proteins.
- Sample requirement: ≥200μg/protein, concentration ≥0.5μg/μL.
Advantages of the protein sequencing service
- One-stop service: Customize exclusive solutions for you according to your testing purpose and budget.
- Professional technology platform: Based on high resolution (greater than 105), high quality accuracy (less than 1ppm) mass spectrometry platform to ensure the accuracy of identification results.
- Advanced data processing algorithms: Developed the calculation method of mass spectrum raw data. Adopt the principle of wide entry and narrow exit. The protein sequence can be accurately identified without missing any valid data information.
- Wide range of application: no species-specific restrictions, can be applied to all species in theory.
Case: Identification and Characterization of Peptide Alterations in Breast Cancer Cell Lines Using LC-MS/MS Analysis
Background
Breast cancer remains a significant health concern, necessitating advanced techniques for understanding molecular alterations. This study focuses on employing Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) to identify and characterize peptide alterations in breast cancer cell lines (MCF7, SKBR3) and a non-tumorigenic cell line (MCF10). The goal is to assess the validity and biological relevance of observed alterations, shedding light on potential implications for cancer progression and treatment strategies.
Samples
The study utilizes three cell lines—MCF7 and SKBR3 representing breast cancer, and MCF10 as a non-tumorigenic control. These cell lines undergo specific culture conditions and processing, generating three biological replicates for each cell state. Cell extracts, including nuclear and cytoplasmic fractions, are prepared for subsequent LC-MS/MS analysis.
Technical Methods
Cell Culture: Cells cultured as per manufacturer protocols with optimal growth media supplemented with specific factors, and subjected to serum deprivation and re-stimulation.
Cell Extract Preparation: Nuclear and cytoplasmic fractions separated using the Cell Lytic™ NuCLEAR™ kit. Protein extracts quantified and reduced before enzymatic digestion with trypsin.
LC-MS/MS Analysis: Micro-LC 1100 system employed for nano-LC separations using in-house-prepared columns. Mobile phases A and B, eluent gradient, and MS settings specified.
Bioinformatics Analysis: Proteome Discoverer 1.4 used for raw MS data processing. Database searches conducted against canonical and mutated peptide databases (XMAn-v1). Functional classification performed using DAVID 6.8.
Results
The analysis yields comprehensive insights into the peptide alterations observed in the breast cancer cell lines. Key findings include the identification of mutated peptide sequences, assessment of peptide false discovery rates, and the exploration of biological relevance through the mapping of missense mutations to cancer driver genes. The study provides valuable information for understanding the molecular landscape of breast cancer, contributing to ongoing efforts in cancer research and personalized medicine.
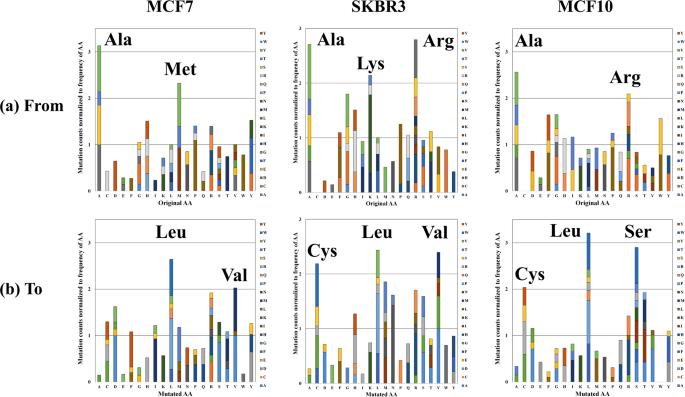 Stacked column charts depicting the relative frequency of amino acid mutations.
Stacked column charts depicting the relative frequency of amino acid mutations.
Reference
- Lazar, Iulia M., et al. "Proteogenomic analysis of protein sequence alterations in breast cancer cells." Scientific Reports 9.1 (2019): 10381.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.