Membrane Proteomics Services
Online InquiryThe membrane is the cell's barrier to the outside world and the interface between the cell's internal and external environment. It isolates cells from the surrounding environment and maintains a relatively stable intracellular environment. Cell surface proteins (membrane proteins) perform important functions such as material exchange inside and outside the cell, cell recognition and immune response, signal transduction and regulation, and energy transfer. To better understand these processes, it is necessary to reveal the identity of membrane proteins and monitor the modification and regulation of their expression. In addition, many drugs currently target membrane proteins. Research on membrane proteins can help develop new drug targets and new treatments.
Creative Proteomics provides a one-stop service for membrane proteomics, which aims to conduct a comprehensive analysis of membrane from a holistic perspective. The main contents include: 1) Enrichment of cytoplasmic membrane. Density gradient centrifugation, antibody immunomagnetic bead purification, and biotin labeling can be used to enrich the cytoplasmic membrane; 2) extraction, separation, and identification of cytoplasmic membrane proteins. Shotgun and top-down strategies are optional. 3) Quantitative study of membrane proteome. We can provide labeled and labeled-free protein quantification methods and complete verification services; 4) Data analysis and bioinformatics analysis.
We Provide the Following Services, including but not Limited to:
Enrichment of cell membrane
- Density gradient centrifugation
- Antibody immunomagnetic bead purification: the antibody immobilized on magnetic beads is used to perform antigen-antibody reaction with cell surface proteins to enrich the membrane.
- Biotin labeling method: Both avidins and streptavidins have high affinities to biotin and can be combined into solid, irreversible complexes。With harsh cleaning conditions, non-specifically bound components can be washed away. In particular, by washing with a high-salt and high-alkali solution, cytosolic proteins and membrane-associated proteins can be effectively removed, and a high-purity membrane can be obtained.
Extraction, isolation and identification of membrane proteins
- Extraction of membrane proteins: We use organic solvents to dissolve membrane proteins. Organic solvent systems do not inhibit the ionization efficiency of mass spectrometry and have no effect on liquid phase separation. No special treatment is required to remove them, which reduces the loss of low-abundance proteins.
- Isolation and identification of membrane proteins: We have two strategies, including shotgun and top-down. The shotgun strategy is to directly digest membrane protein samples. After digestion, the complex peptide mixture was further separated by liquid chromatography and identified by mass spectrometry according to the characteristics of peptide charge and hydrophobicity. The top-down strategy does not require enzyme digestion, and is directly identified by mass spectrometry. This method is more suitable for identifying protein post-translational modifications.
Quantitative analysis of membrane proteins
Both labeled and labeled-free quantification strategies are available. Label-based quantification technologies include: SILAC, TMT, and iTRAQ. Labeled-free quantitative technologies include SRM/MRM, PRM and SWATH technologies. Different quantitative techniques have their unique advantages and characteristics.
Bioinformatics analysis of membrane proteins
Conventional analysis includes: isoelectric point, relative molecular mass, enzymatic cutting characteristics, hydrophobicity, charge distribution, prediction of transmembrane region and transmembrane direction, etc. In addition, there is also the bioinformatics analysis of membrane proteins involved in the identification and quantification of membrane proteins. See the table below for details of "Data analysis of plasma proteomics".
Advantages of the Membrane Proteomics Service:
- One-stop service: We will customize a solution for you based on your samples and needs. You can also choose which technique to use for analysis.
- High performance mass spectrometry system: equipped with Q Exactive Hybrid Quadrupole-Orbitrap and Agilent 6540 Q-TOF high performance mass spectrometer with multiple mass spectrometry techniques such as SRM, PRM and SWATH full scan collection, which effectively improving the identification and quantification of low abundance subcellular related proteins.
- Multi-database integration: by integrating multiple related high-quality protein databases, the accuracy and flux of subcellular protein localization can be greatly improved.
- Multi-dimensional data analysis: in addition to the comparison with the more complete related subcellular database, multiple software was used for the prediction analysis of subcellular localization based on the calculation method to further increase the identification flux.
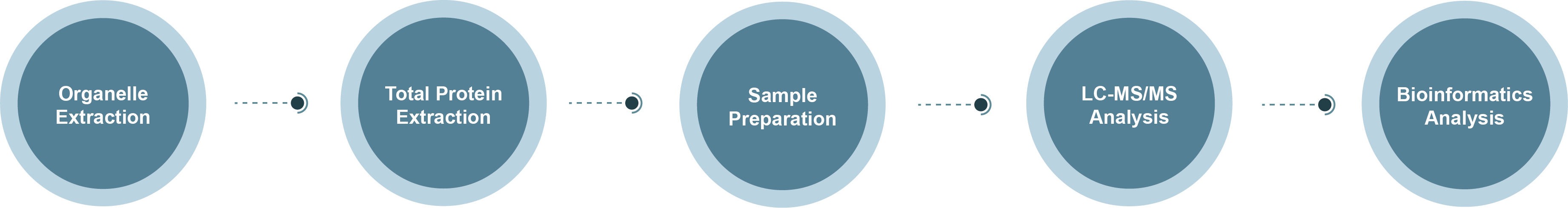
Membrane Proteomics Technology Roadmap
Data Analysis of Membrane Proteomics
| 1. Data output statistics and quality control | 2. Protein identification analysis (Density map, Scatter plot, Venn diagram) | 3. Quantitative protein analysis (Density map, Scatter plot, Venn diagram) |
| 4. Protein GO analysis | 5. Protein pathway analysis | 6. Protein COG analysis |
| 7. GO enrichment analysis of differential proteins | 8. Pathway enrichment analysis of differential proteins | 9. COG analysis of differential proteins |
References
- Cordwell S J, Thingholm T E. Technologies for plasma membrane proteomics. Proteomics, 2010, 10(4): 611-627.
- Mock H P, Matros A, Witzel K. Plant Membrane Proteomics. Springer, 2018.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.