Mitochondrial Protein Phosphorylation Analysis
Online InquiryMitochondria are essential organelles that play a major role in energy metabolism, cell survival and other critical biological processes. Mitochondrial dysfunction has been found to be implicated in many human diseases, which often manifest in a tissue-specific manner. Thus, mitochondrial adaptation relies on a multifunctional proteome dedicated to meeting the specific needs of different tissues. Protein phosphorylation is one of the most well-known post-translational modifications (PTMs) that occur in all domains of life. Increasing evidence suggests that phosphorylation may also play an important role in the regulation of tissue-specific mitochondrial function and pathophysiology.
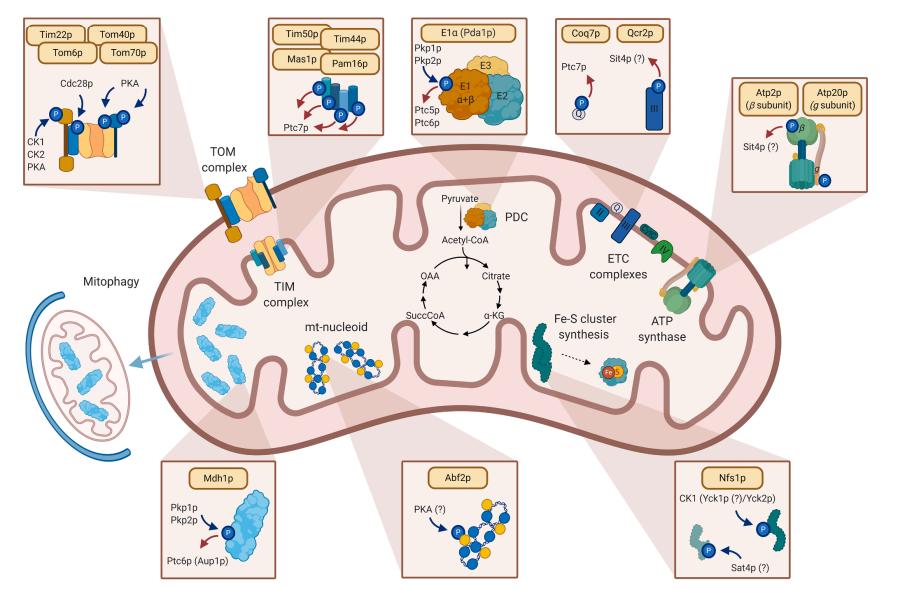 Fig. 1 Highlights of mitochondrial processes affected by protein phosphorylation. (Frankovsky, Jan, et al, 2021)
Fig. 1 Highlights of mitochondrial processes affected by protein phosphorylation. (Frankovsky, Jan, et al, 2021)
Overview of mitochondrial protein phosphorylation
One of the oldest known signaling mechanisms for mitochondrial regulation of metabolism is the targeting of metabolic proteins for reversible phosphorylation. Mitochondrial ATP is generated by oxidative phosphorylation (OXPHOS) via the components of the electron transport chain (ETC), a process regulated by reversible phosphorylation. Mitochondrial protein phosphorylation also mediates cell death signals. In addition to the upregulation of mitochondrial outer membrane protein phosphorylation to induce apoptosis, phosphorylation signaling cascades have also been found to be involved in the control of mitochondrial permeability transition pore (mPTP)-mediated apoptosis. Furthermore, the phosphorylation of proteins in the mitochondria is closely associated with several diseases. STAT3 is a mitochondrial protein involved in cellular respiration, and its phosphorylation has been shown to be important in maintaining ETC complex I (CI) and complex II (CII) respiratory activity.
Mitochondrial protein phosphorylation analysis at Creative Proteomics
Based on cell fractionation and biochemical methods combined with high-throughput techniques of mass spectrometry (MS) techniques, Creative Proteomics is proud to offer mitochondrial protein phosphorylation analysis services. Our team of experts focuses on various organisms or mitochondria/phosphopeptide enrichment protocols, improved analysis pipelines, and multiple mitochondrial protein annotation databases. We are committed to precisely quantifying different protein expression and phosphorylation patterns in mitochondria in various cells and tissues. Our services enable in-depth measurement of phosphoproteomes, providing unbiased insights into mitochondrial composition and function.
Workflow of our service
- Experimental consultation and design
- Sample preparation and isolation of ultra-pure mitochondria
- Mitochondrial phosphoproteome sample preparation
- LC-MS/MS analysis
- Raw data analysis
- Bioinformatics analysis
Sample requirements
- Acceptable samples:
-Cell samples (cell volume > 107)
-Tissue samples (animal tissue >2000 mg, plant tissue >1000 mg)
-Body fluid samples (blood, urine)
-Microorganisms dry weight > 200 mg
- Sample shipping:
Sufficient amount of dry ice for shipping, or consult our technical staff before sending samples
- Before the formal experiment, we will always test the samples you provide
Stable isotope labeling with amino acids in cell culture (SILAC) and isobaric tags for relative and absolute quantification (iTRAQ) are currently the most commonly used techniques in MS-based quantitative phosphoproteomics. In addition to a large-scale discovery phosphoproteomics approach, we also apply multiple reaction monitoring (MRM) to quantify targeted phosphopeptides. If you are interested in our services, please don't hesitate to contact us.
Reference
- Frankovsky, Jan, et al. "Mitochondrial protein phosphorylation in yeast revisited." Mitochondrion 57 (2021): 148-162.
* For Research Use Only. Not for use in diagnostic procedures.