Exosome Proteomics in Disease Mechanisms Analysis
Online InquiryExosomes,as small extracellular vesicles,have an critical role in intercellular communication. Exosomes are released by various cell types and carry various of substances such as proteins, nucleic acids and lipids. With the development of proteomics research, the study of exosomal proteomics is helping scientists to widely used with the study of disease mechanisms and plays an important role in revealing the complexity of various diseases.
Exosome Proteomics in Disease Mechanisms Analysis
Exosome proteomics is essential for understanding the complex processes that underlie many illnesses. Researchers can learn more about the onset, course, and therapeutic effects of disease by examining the protein content of exosomes. Exosome proteomics has a wide range of useful applications in the study of disease processes.
Biomarker Discovery and Early Detection
The discovery of illness-specific biomarkers that can aid in early disease identification and surveillance by exosome proteomics has enormous potential. Exosomes contain disease-associated proteins that can act as early warning signs of pathological diseases before any clinical symptoms appear. The proteome patterns of exosomes from sick and healthy people can be compared to identify possible biomarkers with high sensitivity and specificity for early diagnosis.
Understanding Disease Pathways and Mechanisms
Exosome proteomics research and development allow for a thorough examination of disease processes. Researchers can use exosomal protein carriers to characterize proteins engaged in crucial signaling cascades or significant physiological activities. The comprehension of diverse illness progressions, the connection between cellular communication and disease incidence, and the interactions between cells under various disease situations can all be improved with the aid of this information. For example, exosome proteomics has revealed insights into the molecular events underlying cancer metastasis and the dysregulation of neurodegenerative disease-associated pathways.
Prediction of Disease Progression and Treatment Responses
Analyzing exosomal proteins can aid in predicting disease progression and assessing treatment responses. Changes in the exosomal proteome over time can provide valuable information about disease dynamics and response to therapeutic interventions. Monitoring the abundance of specific proteins within exosomes may offer prognostic insights, enabling clinicians to tailor treatment strategies based on individual patient profiles. Exosome proteomics has shown potential in predicting therapeutic responses in cancer patients undergoing targeted therapies.
Uncovering Mechanisms of Intercellular Communication
Exosomes serve as carriers of bioactive molecules, enabling intercellular communication in both physiological and pathological contexts. Exosome proteomics unravels the cargo of proteins involved in cell-to-cell signaling, immune modulation, and tissue homeostasis. Understanding how disease-associated proteins are packaged and transferred through exosomes can illuminate how cells communicate under normal and diseased conditions. This knowledge has implications for developing interventions that target intercellular communication networks.
Learn more
Methods Used in Exosome Proteomics for Disease Analysis
Liquid Chromatography-Mass Spectrometry (LC-MS)
LC-MS is a cornerstone technique in exosome proteomics. It enables the identification, quantification, and profiling of proteins within exosomes. LC separates proteins based on their physicochemical properties, and MS detects and quantifies the separated proteins. This method allows researchers to analyze complex mixtures of exosomal proteins with high sensitivity and resolution. Advanced LC-MS platforms coupled with data analysis tools facilitate the identification of disease-specific protein signatures, contributing to biomarker discovery and pathway elucidation.
Isolation Techniques
Effective isolation of exosomes is a prerequisite for accurate proteomic analysis. Two commonly employed methods for exosome isolation are ultracentrifugation and size-exclusion chromatography. Ultracentrifugation involves differential centrifugation steps to pellet exosomes from biological fluids. Size-exclusion chromatography separates exosomes based on their size, allowing for the enrichment of exosome-containing fractions. These isolation techniques ensure the purity of exosome samples, minimizing contamination and enabling reliable proteomic analysis.
Immunoprecipitation and Affinity-based Enrichment
Immunoprecipitation (IP) and affinity-based enrichment methods target specific proteins or protein complexes within exosomes. Antibodies against known disease-related proteins can be used to selectively enrich relevant exosomal proteins. This approach is particularly valuable when investigating specific pathways or protein-protein interactions implicated in disease mechanisms. IP coupled with subsequent proteomic analysis provides insights into disease-associated protein networks.
Label-Free Quantitative Proteomics
Label-free quantitative proteomics is an emerging approach that allows for the comparison of protein abundance between different exosome samples. This method relies on spectral counting or intensity-based quantification to measure the relative abundance of proteins. Label-free techniques enable the identification of differentially expressed proteins in disease versus control exosomes, highlighting potential markers or pathways relevant to pathological conditions.
Bioinformatics and Systems Biology Analysis
In conjunction with experimental techniques, bioinformatics and systems biology analyses are essential for interpreting complex proteomic datasets. Data analysis tools facilitate protein identification, quantification, and functional annotation. Pathway enrichment analysis and protein interaction network mapping aid in unraveling disease-related pathways and protein interactions, providing a holistic view of disease mechanisms.
Case study: Exosome Proteomics in Disease Mechanisms Analysis
Background
Exosomes, small extracellular vesicles released by cells, have gained significant attention in recent years for their potential as valuable biomarkers and key players in intercellular communication. These nanosized vesicles carry a cargo of proteins, nucleic acids, and lipids that can shed light on various aspects of cellular health and disease mechanisms.
Sample Selection and Characterization:
A critical aspect of exosome proteomics is the selection and characterization of samples. Researchers often collect exosomes from various biological fluids, such as blood, urine, or cerebrospinal fluid, based on their potential relevance to the disease of interest. For instance, in the study by Thompson et al., cerebrospinal fluid (CSF) samples were collected from individuals with amyotrophic lateral sclerosis (ALS) to investigate proteomic changes in CSF extracellular vesicles (EVs) (Thompson, et al,. 2020). Similarly, Waldron et al. focused on plasma samples to identify biomarkers associated with alcoholic acute pancreatitis (Waldron, et al,. 2018).
Characterization techniques like transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and Western blotting help confirm the presence of exosomes based on their size, morphology, and marker proteins. This initial step ensures that the subsequent proteomic analysis is conducted on genuine exosome populations.
Technical Approach
Proteomic Analysis of Exosomes:
Proteomic analysis is at the heart of exosome research. It involves identifying and quantifying the proteins within exosomes to gain insights into disease mechanisms. Mass spectrometry (MS) is a cornerstone technique for this purpose. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is commonly used to analyze the protein content of exosomes.
Researchers employ advanced LC-MS/MS technology to achieve high-resolution proteomic analysis. This technique allows for the identification of a wide range of proteins present in exosomes, enabling a comprehensive view of the molecular landscape. Through this approach, novel plasma biomarkers were identified in alcoholic acute pancreatitis, shedding light on the pathobiological pathways associated with the disease (Waldron, et al,. 2018).
Pathway Analysis and Functional Insights:
Once the proteomic data is obtained, researchers often perform pathway analysis to gain a deeper understanding of the biological processes implicated in the disease. Researchers utilizes sophisticated bioinformatics tools to unravel complex networks of protein interactions.
In the ALS study by Thompson et al., altered protein homeostasis was observed in CSF EVs, suggesting potential dysregulation in disease-related pathways (Thompson, et al,. 2020). This type of analysis can provide crucial clues about the underlying mechanisms and guide further investigations.
Results
The results of exosome proteomics studies have far-reaching implications in understanding disease mechanisms. In the case of ALS, the identification of specific proteins and altered pathways in CSF EVs provides valuable insights into the pathogenesis of this devastating neurodegenerative disease. These findings may pave the way for the development of diagnostic tools or therapeutic targets (Thompson, et al,. 2020).
In alcoholic acute pancreatitis, the discovery of novel plasma biomarkers opens new avenues for early diagnosis and disease monitoring. Understanding the pathobiological pathways associated with this condition can lead to more effective treatments and improved patient outcomes (Waldron, et al,. 2018).
In conclusion, exosome proteomics is a powerful tool that researchers worldwide are using to uncover the molecular secrets of diseases. Through careful sample selection, advanced proteomic techniques, and in-depth pathway analysis, we are gaining a deeper understanding of disease mechanisms and moving closer to innovative diagnostic and therapeutic solutions.
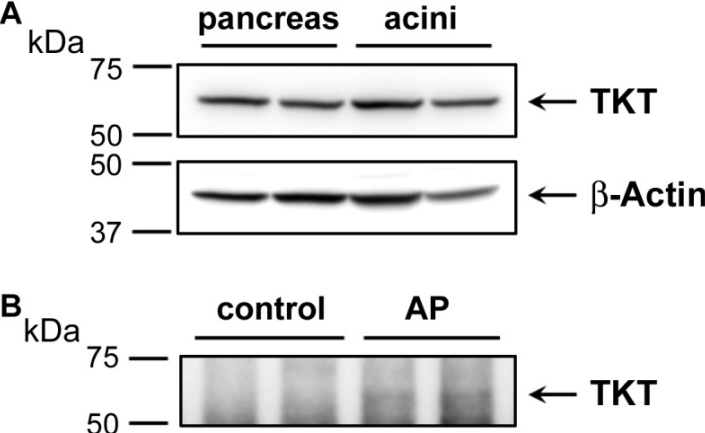 Western blot analysis depicting the expression of transketolase (TKT) in human pancreas and acinar cell samples (Waldron RT, et al,. 2018)
Western blot analysis depicting the expression of transketolase (TKT) in human pancreas and acinar cell samples (Waldron RT, et al,. 2018)
References
- Thompson AG, Gray E, Mäger I, et al. CSF extracellular vesicle proteomics demonstrates altered protein homeostasis in amyotrophic lateral sclerosis. Clin Proteomics. 2020 ;17:31.
- Waldron RT, Lugea A, Gulla A, et al. Proteomic Identification of Novel Plasma Biomarkers and Pathobiologic Pathways in Alcoholic Acute Pancreatitis. Front Physiol. 2018 ;9:1215.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.