Proteomics Solutions of Microbial Pathogens
Despite significant advances in the treatment and prevention of bacterial infectious diseases, they remain a significant threat to human health because of the enormous global burden of morbidity and mortality. Drug-resistant pathogens are on the rise, which greatly reduces our antibiotic pool for treating bacterial infections. Understanding the physiology and pathogenicity of bacteria is essential for vaccine discovery, diagnosis and drug development[1].
MS-based proteomics has made progress in microbiology studies, including protein identification, bacterial proteome quantification, translational protein modification (PTMs) analysis, pathogen-host cell interaction discovery, antimicrobial resistance, and biomarkers. We use state-of-the-art mass spectrometry (phosphate) proteomics techniques to study the pathogenicity mechanisms and characterize the interactions between molecules that mediate host-pathogen interactions comprehensively. Furthermore, complete molecular knowledge was necessary to provide suitable strategies for fighting human microbial pathogens.
MS-based proteomics can be divided into two main categories: discovery and targeted proteomics. During recent years, discovery and targeted proteomics applications with pathogenic bacteria have significantly expanded due to its specificity, reproducibility, and multiplexing capability compared to immunological assays.
Targeted Acquisition Methods
Targeted methods show the highest accuracy and reproducibility among all mass spectrometry methods. Although targeted methods are often used to quantify specific proteins, this technique cannot be practically extended to thousands of proteins thus is limited. Targeted acquisition methods in proteomics can be roughly divided into two main approaches: selected reaction monitoring (SRM), also known as multiple reaction monitoring (MRM) and parallel reaction monitoring (PRM). Our Targeted Proteomics Services typically quantify up to 150 proteins with high precision and throughput in 6 orders of magnitude dynamic range.
Discovery Proteomics Strategy
Discovery proteomics aims to comprehensively identify proteins in a sample, especially in samples where bacterial pathogens need to be studied at low abundancy and complex matrices, such as blood. DDA analysis is hampered by its lower dynamic range, while DIA(Data independent acquisition) MS approaches are the methods of choice to overcome this problem. For example, SWAT-MS, a DIA strategy that combines the advantages of both DDA and targeted proteomic strategy, allows for the systematic and reproducible analysis of thousands of proteomes under different conditions and samples. Thus, DIA MS is more suitable for quantitative clinical proteomics.
We offer DIA MS services for large-scale clinical samples quantitative analysis that can identify up to 9000 proteins at a time.
Applications
Monitoring bacterial metabolic processes
Bacterial pathogenicity mechanisms research
Antibiotic-resistance mechanisms study
Biomarker discovery and diagnostic applications
Monitoring food safety
Workflow
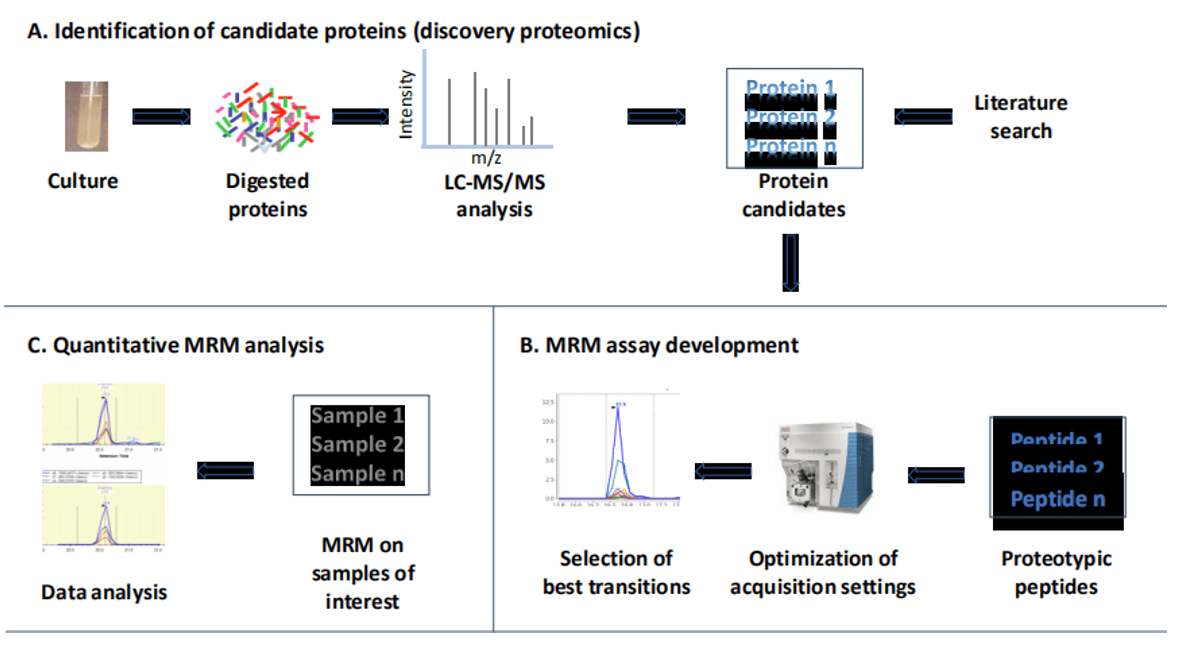 Schematic representation of a typical workflow to study bacteria proteomes[1]
Schematic representation of a typical workflow to study bacteria proteomes[1]
Advantages
High accuracy
High throughput, more than 9000 proteins can be identified at once
Quantitatively identify nearly all detectable molecules, covering low-abundance proteins/peptides
High repetition rate
Complete and comprehensive sample information storage in the first analysis
Sample Requirements
Animal and clinical tissue specimens: 200 mg/sample
Cells, microorganisms: 1×107 cells/sample
Plant tender leaves and buds: 500 mg/sample
Plant seeds, fruits: 100 mg/sample
Report
- Experimental steps
- Relevant parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
References:
- Sara Saleh et al. Targeted proteomics for studying pathogenic bacteria. Proteomics 2019 Aug;19(16):e1800435.
- Nelson C. Soares et al. Editorial: Proteomics of Microbial Human Pathogens. Microbiol., 04 2016.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 Schematic representation of a typical workflow to study bacteria proteomes[1]
Schematic representation of a typical workflow to study bacteria proteomes[1]
 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)