DIA Quantitative Proteomics Service
In recent years, a new mass spectrometry data acquisition method, data-independent acquisition (DIA) mode, has been developed. It is an improved technique to the traditional shotgun analysis (data-dependent acquisition), which enables unbiased MS analysis of all signals in a complex sample, resulting in higher peptide coverage. The DIA scan mode divides the entire full scan range of the mass spectrum into several windows, and selects, fragments, and detects all ions in each window at high speed and cyclically, so as to obtain fragment information of all ions in the sample without omission and difference.
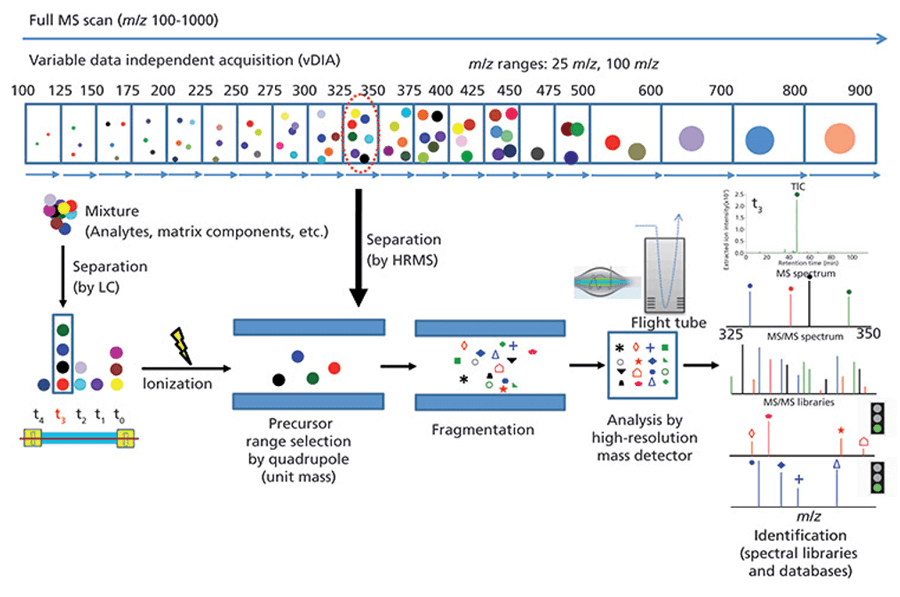 DIA workflow (LCGC North America.Volume 35, Issue 10, pg 756–759)
DIA workflow (LCGC North America.Volume 35, Issue 10, pg 756–759)
Our DIA Quantitative Proteomics Service
Our DIA quantitative proteomics service uses the AB SCIEX Triple-TOF 5600 plus high-resolution mass spectrometry system, which has a high resolution, accurate mass number stability, high sensitivity and high scan speed. The main DIA technologies we use are SWATH-MS, SRM™, MSX-DIA, PCT-DIA and GPF-DIA. All you need to do is send us your samples and we will take care of the following, including protein extraction, proteolysis, peptide separation, mass spectrometry analysis, raw mass spectrometry data analysis, and bioinformatics analysis.
Advantages of Orbitrap-based DIA technology:
(1) High qualitative and quantitative accuracy, typically acquired at 35,000-60,000 resolution.
(2) Higher data throughput with a stable mass axis at 1 ppm over time, without internal standard correction or frequent external standard correction.
(3) Ultra-high sensitivity and high spectral quality, amol (10-18 mol) level.
(4) High throughput, capable of monitoring all target proteins simultaneously.
(5) Ultra-fast scanning speed, 12-20 Hz.
(6) Dynamic range and linear range of more than 5 orders of magnitude.
(7) Unique msxDIA to further improve selectivity.
Workflow:
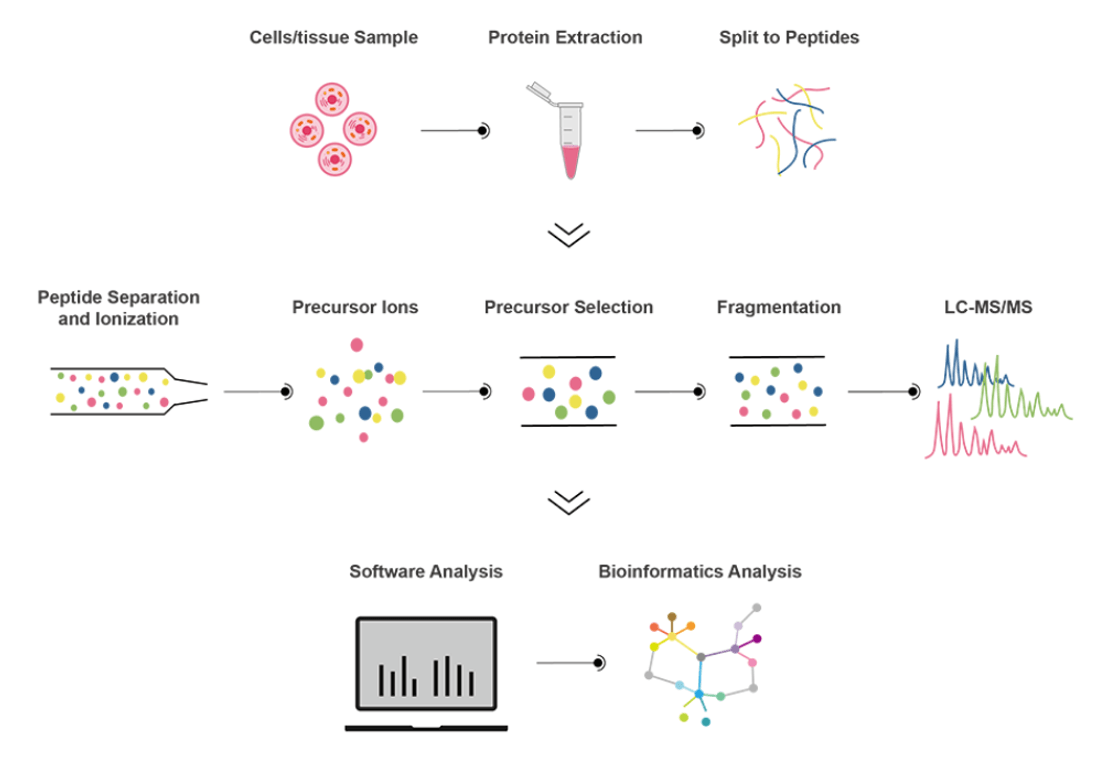
Data Analysis
| Standard data analysis content |
| Statistical analysis of identification results | Sample protein and peptide identification histogram, PCA distribution map, quantitative variance statistical analysis |
| Bioinformatics analysis | Differential up- and down-regulated protein KEGG map, KEGG pathway function attribution, etc. |
| GO and G0 enrichment, KEGG and KEGG enrichment analysis, PPI interaction network and module analysis |
| Advanced data analysis content |
| Biomarker screening | Integrated machine learning, LASSO regression analysis, marker panels, ROC analysis |
| Molecular typing analysis | Unsupervised cluster typing analysis, proteome + transcriptome analysis, kinase analysis |
| Joint analysis of clinical characterization and omics results |
| Disease-related functional module analysis | WGCNA co-expression analysis |
| Survival curve analysis | Clinical typing and survival curve analysis |
Sample Requirements:
- Animal and clinical tissue specimens: 200mg/sample
- Serum, plasma: 200μL/sample
- Cells, microorganisms: 1×107cells/sample
- Plant tender leaves and buds: 500mg/sample
- Plant seeds, fruits: 100mg/sample
Detection Platforms:

Applications:
Clinical and Pharmacology Applications:
- Research on disease biomarkers.
- Research on molecular mechanism of disease development.
- Research on chemical or biological drug targets.
- Research on novel viral inhibitors.
- Research on signal transduction of chemical/biological drug mechanism of action.
Agriculture and Forestry Applications:
- Animal husbandry: Improvement of meat quality and breeding conditions.
- Agriculture: Research on the development mechanisms of agricultural crops.
- Food science: Research to improve food safety, taste and nutrition.
Report:
- Experimental steps
- Relevant parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 DIA workflow (LCGC North America.Volume 35, Issue 10, pg 756–759)
DIA workflow (LCGC North America.Volume 35, Issue 10, pg 756–759)


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)