4D-Methylation Proteomics Services
Protein methylation is a universal modification that participates in various processes, such as gene expression regulation and interaction. Protein methylation is activated by the enzymatic transfer of methyl to an amino acid residue, usually on arginine and lysine, sometimes histidine, cysteine, asparagine, etc. Statistically, up to three methyl groups can be attached simultaneously, with a corresponding increase in molecular weight of 14.02 Da/28.03 Da/42.05 Da for mono/bi/tri-methylation. Mass spectrometry can precisely determine the molecular weight changes and identify the methylation-modified peptides and sites.
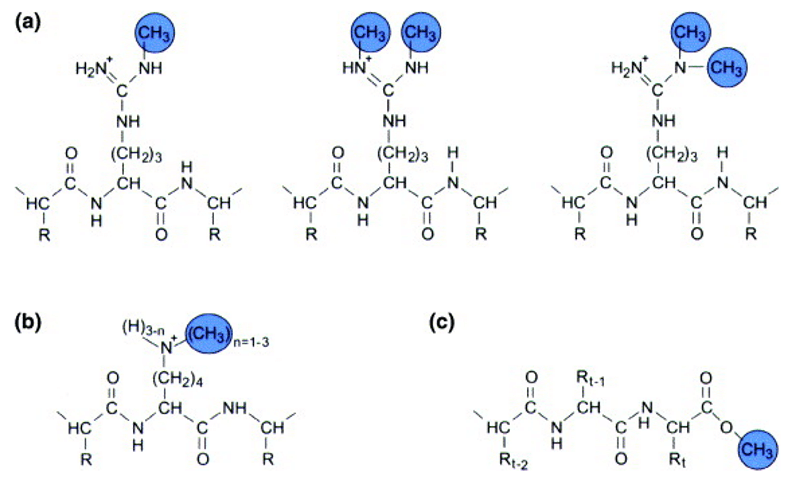 The methylation of amino acid constituents of proteins (John M. Aletta et al. 1998)
The methylation of amino acid constituents of proteins (John M. Aletta et al. 1998)
Based on the heterogeneity and low relative abundance of post-translational protein modifications, the accurate quantification of PTMs confronts various challenges. On the one hand, the low abundance of post-translational modifications requires more advanced sensitivity and identification accuracy. On the other hand, different modification sites occur within the peptide resulting in tautomeric modified peptides, and limiting the traditional mass spectrometry platform to perform separation for these co-eluted tautomeric peptides.
The introduction of ion trickle separation concept has brought proteomics into a new 4D era. The novel generation of 4D proteomics technology adds a fourth dimension, ion mobility, to traditional proteomics and significantly improves analytical performance.
Our 4D-Methylation proteomics Services
Creative proteomics' 4D-label free methylation proteomics is based on the new generation timsTOF Pro mass spectrometer for methylation differential proteomics analysis. We provide "one-stop" proteomics services, from a large-scale screening of PTMs proteomics, PRM validation of targeted proteomics/PTMs omics, to high-quality antibody-assisted validation and subsequent molecular mechanism studies.
Workflow
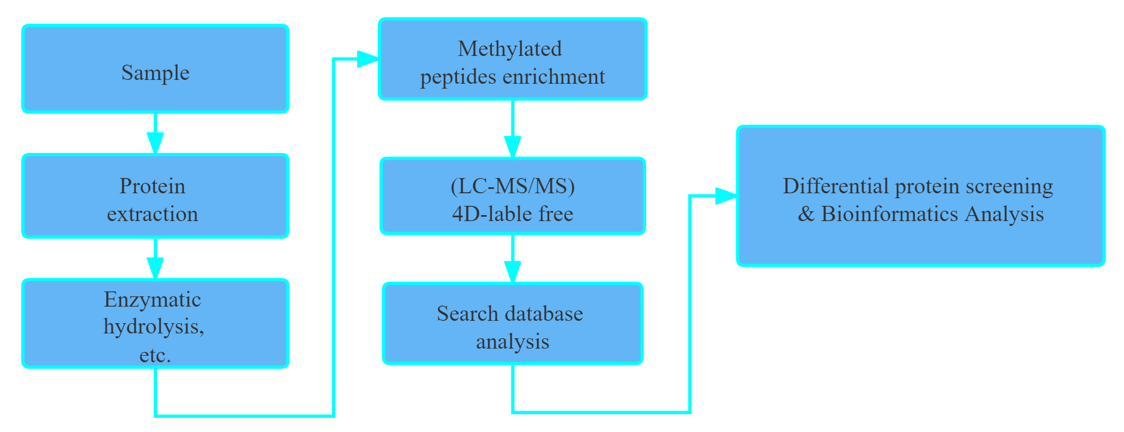
Applications
- Chromatin structure as well as transcriptional regulation.
- Maintenance of protein stability, regulation of protein transcription, binding activity, DNA replication, and damages.
- Closely related to diseases such as cancer, aging, Alzheimer's disease, and epigenetics research content.
Advantages
- Super performance substantially improve the sensitivity and depth of modification histology analysis.
- Ion mobility separation solves isomerism-related problem and provides more reliable quantitative modification results.
Data Analysis
| Standard Data Analysis Content |
| Mass spectrometry data analysis | Spectral peptide quality deviation distribution, peptide length distribution, unique peptide number distribution, protein coverage distribution |
| Protein Expression Analysis | Protein abundance value distribution of PTMs, protein abundance ratio distribution of PTMs between samples, PCA analysis, statistical analysis of significant differences |
| Protein Functional Analysis | Total protein and differential protein GO secondary classification, COG function classification, KEGG annotation, subcellular organelle location, domain annotation, signal peptide prediction and PPI prediction; differential protein GO, KEGG, domain enrichment analysis |
| Advanced Data Analysis Content |
| WGCNA Analysis | Predict functional clustering and network interactions by protein expression level; correlate with phenotype data to obtain key proteins or protein complexes that influence phenotype |
| Trend Cluster Analysis | Obtain protein expression trend patterns |
Report
- Experimental steps
- Relevant experimental parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 The methylation of amino acid constituents of proteins (John M. Aletta et al. 1998)
The methylation of amino acid constituents of proteins (John M. Aletta et al. 1998)

 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)