4D-Ubiquitination Proteomics Services
Protein ubiquitination is often accompanied by ubiquitin-proteasome degradation, leading to changes in protein stability. Protein ubiquitination also participated in various bioregulatory processes, such as proliferation, apoptosis, endocytosis, and immunity. Mechanistically, protein ubiquitination goes through the triple enzyme cascade (E1-E2-E3) reaction that allows single or multiple ubiquitin molecules to attach to amino acid (Lys) side chains and is often accompanied by proteasomal degradation, which regulates protein expression levels changes. The ubiquitination-modified lysine covalently links the ubiquitin molecule, i.e., two glycine K-ε-GG residues, resulting in a mass shift of 114.1 Da. Mass spectrometry can precisely determine the mass shift of 114.1 Da in molecular weight to detect the ubiquitination-modified peptides and sites.
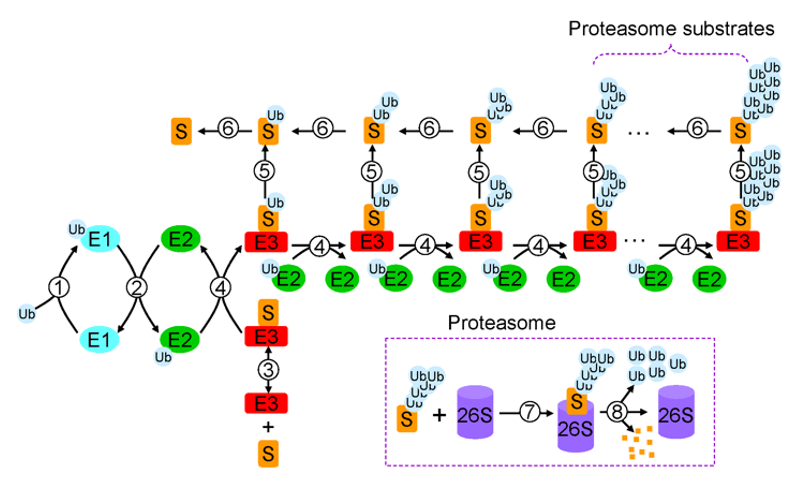 Ubiquitination (Xu L,2012)
Ubiquitination (Xu L,2012)
Our 4D-Ubiquitination Proteomics Services
Creative proteomics uses the 4D proteomics technology to provide you with protein ubiquitination services. 4D label-free ubiquitinated proteomics technology is based on a new generation of timsTOF Pro mass spectrometer to perform ubiquitination differential proteomics analysis. 4D- label label-free PTMs Omics can effectively distinguish modified isomeric peptides to significantly improve identification depth and achieve comprehensive improvement of proteomics in terms of coverage depth, sensitivity, and throughput with less loading volume and faster scanning speed.
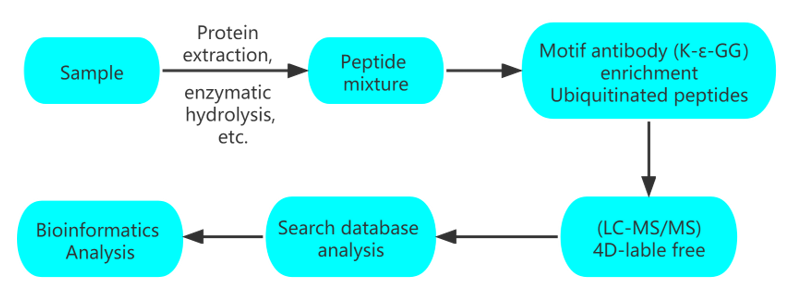
Workflow
Applications
- Neurological and degenerative diseases research.
- Tumor pathogenesis research.
- Pharmacological and pharmacological target studies.
Data Analysis
| Standard Data Analysis Content |
| Mass spectrometry data analysis | Spectral peptide quality deviation distribution, peptide length distribution, unique peptide number distribution, protein coverage distribution |
| Protein Expression Analysis | Protein abundance value distribution of PTMs, protein abundance ratio distribution of PTMs between samples, PCA analysis, statistical analysis of significant differences |
| Protein Functional Analysis | Total protein and differential protein GO secondary classification, COG function classification, KEGG annotation, subcellular organelle location, domain annotation, signal peptide prediction and PPI prediction; differential protein GO, KEGG, domain enrichment analysis |
| Advanced Data Analysis Content |
| Protein Gene Chromosome Localization | Obtain genes encoding proteins distribution on chromosomes |
| WGCNA Analysis | Predict functional clustering and network interactions by protein expression level; correlate with phenotype data to obtain key proteins or protein complexes that influence phenotype |
| Trend Cluster Analysis | Obtain protein expression trend patterns |
| Molecular Typing | Molecular typing for large cohort samples |
| Survival Curve Analysis | Study the relationship between influencing factors, survival time, and outcome |
| ROC Curves | Evaluate predictive accuracy by combining specificity and sensitivity, such as biomarker impact assessment on tumor grade |
Technical Advantages
- The support of 4D technology improves quantification accuracy and identification depth, making it more suitable for trace samples;
- Flexible and convenient;
- Cutting-edge technology-based on a new generation of mass spectrometry platform (Bruker timsTOF pro), nearly doubled detection speed;
- Rich experience - over 10 years of protein platform service experience
- Data assurance - gold standard antibodies and strictly controlled modification enrichment operation to ensure stable and reliable data results.
- System solutions - provide modified omics for verification, upstream and downstream joint analysis, and to achieve a fast "one-stop" service.
Sample Requirements
We can quickly extract proteins from various samples and design personalized experimental schemes according to specific experimental purposes based on our unique protein extraction technology. Specific requirements are as follows:
| Sample Type | Protein | # of Cells | Animal Tissue | Plant Tissue | Blood | Urine | Serum | Microbes |
| Quantify | 100 ug | 1×107 cells | 1 g | 200 mg | 1 mL | 2 mL | 0.2-0.5 mL | Dry weighed: 200 mg |
Report
- Experimental steps
- Relevant experimental parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
Recommended Combination
Comprehensive protein expression levels analysis: transcriptomics + proteomics + ubiquitination.
References:
- Genome-wide identification of U-box genes and protein ubiquitination under PEG-induced drought stress in potato. 2021. Physiologia Plantarum.
- Rewiring of the ubiquitinated proteome determines ageing in C. elegans. 2021. Nature.
- Xu L, Qu Z. Roles of protein ubiquitination and degradation kinetics in biological oscillations. PloS one, 2012, 7(4).
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 Ubiquitination (Xu L,2012)
Ubiquitination (Xu L,2012)

 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)