Drug Target Identification Solutions
The field of drug target discovery is becoming popular with great potential to advance biomedical research and chemical genomics. Since many small molecule bioactive molecules exert their functions by interacting with protein targets and an increasing number of drugs exhibit targeting of multiple proteins. Therefore, the identification of target recognition is of great importance for drug development. Drug target identification not only deepens the understanding of drug mechanism of action, but also reveals its potential therapeutic applications and adverse effects.
Revealing the target of a small molecule compound is key to gaining insight into its mechanism of action and potential toxicity. However, considering that the non-specific binding of many small molecules remains a major problem, which may reduce the accuracy of target identification and hinder the drug development process, a high-throughput, high-precision approach is necessary. To overcome these limitations, quantitative proteomics approaches have been developed in recent years, including metabolic labeling, chemical labeling, and label-free methods. These new methods are combined with activity-based protein profiling (ABPP), allowing a rapid process and accurate results.
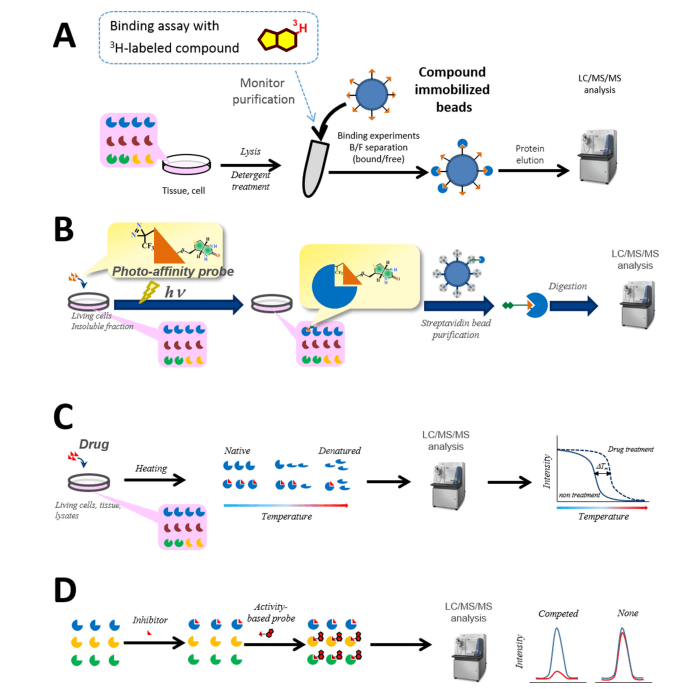 Target deconvolution by chemical proteomics (Kubota, 2016)
Target deconvolution by chemical proteomics (Kubota, 2016)
Drug Target Identification Services
We offer drug target analysis services that combine ABPP with quantitative proteomics to identify natural and traditional drug targets.
Our discovery proteomics platform can quantify thousands of proteins per sample of almost any biological sample, such as cell culture, tissue, or body fluids. This enables the identification of significantly regulated proteins in disease models for therapeutic exploration.
Our DIA MS and 4D DIA technologies are novel, label-free methods that can target and non-target the entire proteome to detect protein changes with peptide-level resolution.
Benefit Fields
Clinical validation of drug candidates
Discovery of the novel targets for lead compounds developed using cell-based phenotypic screens
Drug activity and toxicity research
Discover or validate new protein targets for disease, such as cancer inflammatory, immunology, or other diseases
Workfolw
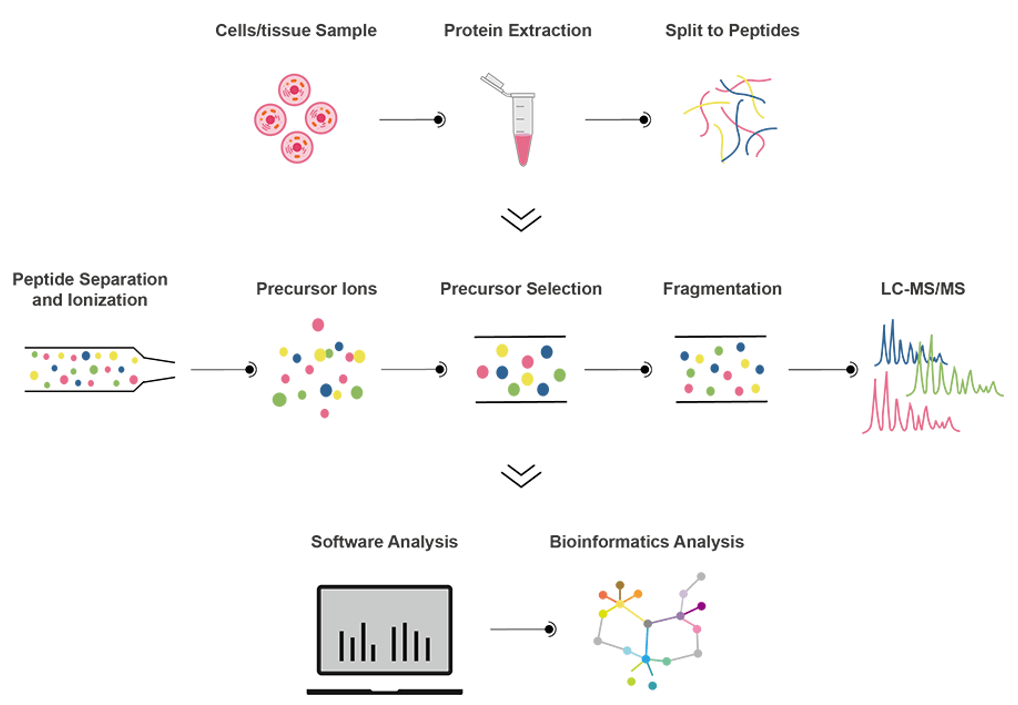
Bioinformatics Analysis
| Problems to be Solved | Bioinformatics Analysis |
| Quality Assessment of Protein | SWATH Data Analysis |
| Protein Comparison of Different Samples | Multivariate PCA Analysis |
| Protein Statistical Analysis | Venn Diagram |
| Volcano Plot |
| Functional Annotation | KEGG Annotation |
| GO Annotation |
| COG Annotation |
| Clustering Analysis | Hierarchical Clustering |
| K-Means Clustering |
| Network Analysis | STRING Analysis |
Analytical Platforms
AB SCIEX Triple-TOF 5600-plus, Q-Exactive, Orbitrap Fusion
Key Features
- High accuracy
- High throughput, more than 9000 proteins can be identified and quantitated at once
- Quantitatively identify nearly all detectable molecules, covering low abundance proteins/peptides
- High repetition rate
- Complete and comprehensive information storage of samples in the first analysis
- High repetition rate
Sample Requirements
Our special protein extraction technology allows us to quickly extract proteins from various samples and design personalized experimental schemes according to different experimental purposes. Specific requirements are as follows:
| Sample Type | Protein | # of Cell | Animal Tissue | Plant Tissue | Blood | Urine | Serum | Microbes |
| Quantify | 100 ug | 1×107 cells | 1 g | 200 mg | 1 mL | 2 mL | 0.2-0.5 mL | Dry weighed: 200 mg |
Note
Please prepare enough dry ice or ice packs to ensure low temperature during sample.
Delivery
Our service is for research use only and is not intended for diagnosis.
Report
- Experimental steps
- Relevant experiment parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
References:
- Kubota, Kazuishi et al. Target deconvolution from phenotype-based drug discovery by using chemical proteomics approaches. Pharmacol Ther. 2016;162:10-22.
- Xiao Chen et al. Target identification with quantitative activity based protein profiling (ABPP). Proteomics. 2017; 17:3-4.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 Target deconvolution by chemical proteomics (Kubota, 2016)
Target deconvolution by chemical proteomics (Kubota, 2016)

 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)