FFPE Quantitative Proteomics Solutions
At present, the majority of current clinical proteomics studies are still carried out on fresh or frozen tissue samples, which often lack sufficiently long follow-up information, limiting their practical applications. On the other hand, Formalin Fixed Paraffin-Embedded (FFPE) has been the most commonly used method of sample preservation for the clinical histopathological diagnosis of diseases. FFPE also is an economical archival choice, as tissues can be stored at a great density at room temperature (RT) for years or decades while maintaining integrity for pathology analyses. An estimated 500 million FFPE cancer tissues are stored to date, and this number is rapidly rising. These present an invaluable resource for studying molecular mechanisms underlying diseases and testing potential biomarkers and discovering new ones. If these samples can be applied to proteomic analysis, they will undoubtedly be of better clinical value. The recent technological progress of MS-based proteomic methods now enables the unbiased and large-scale investigation of proteomes at ever greater depth.
Our FFPE Quantitative Proteomics Service
We apply advanced DIA proteomics analysis technology to FFPE tissue protein extracts, which can reveal a large amount of disease-related functional information, so as to help you identify meaningful biomarkers related to the diagnosis, prognosis, monitoring and treatment of specific diseases. We also offer highly multiplexed targeted proteomics with absolute quantification for customized panels of proteins.
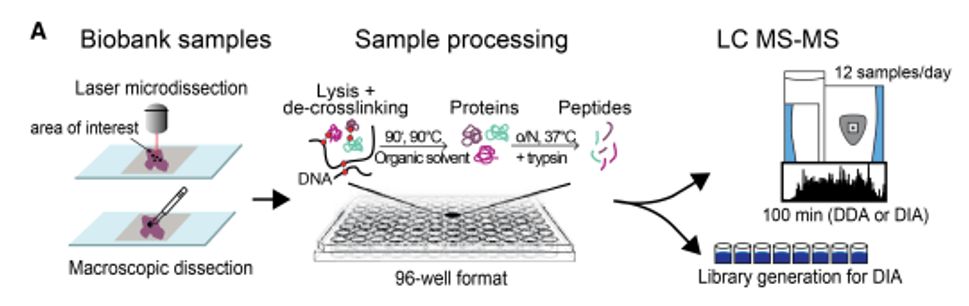 Mass spectrometry-based workflow for FFPE tissue analysis (Fabian Coscia, et al, bioRxio. 2020)
Mass spectrometry-based workflow for FFPE tissue analysis (Fabian Coscia, et al, bioRxio. 2020)
Help You Overcome Technical Difficulties
Tissue Treatments such as formaldehyde fixation and paraffin embedding lead to extensive molecular cross-linking of proteins, nucleic acids and other molecules in tissue cells, making it quite difficult to obtain proteome within FFPE tissues. Formaldehyde fixation limits the extraction efficiency of proteins, reduces the immune response activity, and may lead to unclear identification of proteins by mass spectrometry. Therefore, an efficient method for protein extraction from FFPE tissues is needed. Based on this, Creative Proteomics has developed an extraction method that can obtain high-quality proteins from FFPE samples after extensive literature research and R&D testing.
Data Analysis
| Standard data analysis content |
| Statistical analysis of identification results | Sample protein and peptide identification histogram, PCA distribution map, quantitative variance statistical analysis |
| Bioinformatics analysis | Differential up- and down-regulated protein KEGG map, KEGG pathway function attribution, etc. |
| GO and G0 enrichment, KEGG and KEGG enrichment analysis, PPI interaction network and module analysis |
| Advanced data analysis content |
| Biomarker screening | Integrated machine learning, LASSO regression analysis, marker panels, ROC analysis |
| Molecular typing analysis | Unsupervised cluster typing analysis, proteome + transcriptome analysis, kinase analysis |
| Joint analysis of clinical characterization and omics results |
| Disease-related functional module analysis | WGCNA co-expression analysis |
| Survival curve analysis | Clinical typing and survival curve analysis |
Advantages
- High accuracy: QC CV as low as 25% for large projects
- High throughput, more than 9000 proteins can be identified at once
- Quantitatively identify nearly all detectable molecules, covering low-abundance proteins/peptides
- High repeatability increased by 30% and up to 90%
- Complete and comprehensive sample information storage in the first analysis
- Standard data and advanced data analysis
- Low cost: reduce the cost by more than 3 times
Sample Requirements
| Sample Type | Protein | # of Cell | FFPE slices |
| Quantify | 100 μg | 1×107 cells | 2×5 μm FFPE slices (each 50-100mm2) |
Report
- Experimental steps
- Relevant experiment parameters
- Mass spectrometry spectra
- Raw data
- Proteomics analysis results
Our FFPE Quantitative Proteomics Service is ideal for long-term projects or projects with large sample sets that require accurate and reproducible quantitation. Please contact us for more details.
Reference:
- Fabian Coscia, et al., 2019, A streamlined mass spectrometry-based proteomics workflow for large scale FFPE tissue analysis. bioRxio. 251(1):100-112. 2020
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 Mass spectrometry-based workflow for FFPE tissue analysis (Fabian Coscia, et al, bioRxio. 2020)
Mass spectrometry-based workflow for FFPE tissue analysis (Fabian Coscia, et al, bioRxio. 2020)
 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)