Cancer Biomarker Discovery Solutions
With the development of mass spectrometry, quantitative proteomics has been widely used in cancer mechanism research. A variety of tumor biomarkers identified by quantitative proteomics contribute to early diagnosis, prognosis, and drug resistance analysis. Three types of samples, including cell lines, clinical specimens, and body fluids, are used for quantitative proteomics studies. Clinical specimens and body fluids have been widely used in cancer research. Liquid biopsy is increasingly recognized as a promising method for the non-invasive identification of clinical biomarkers. Many tumor-related biomarkers have been identified in serum, urine, saliva, and exomes. The quantitative proteomics of liquid biopsies faces great challenges due to the high complexity and the wide dynamic range of proteins.
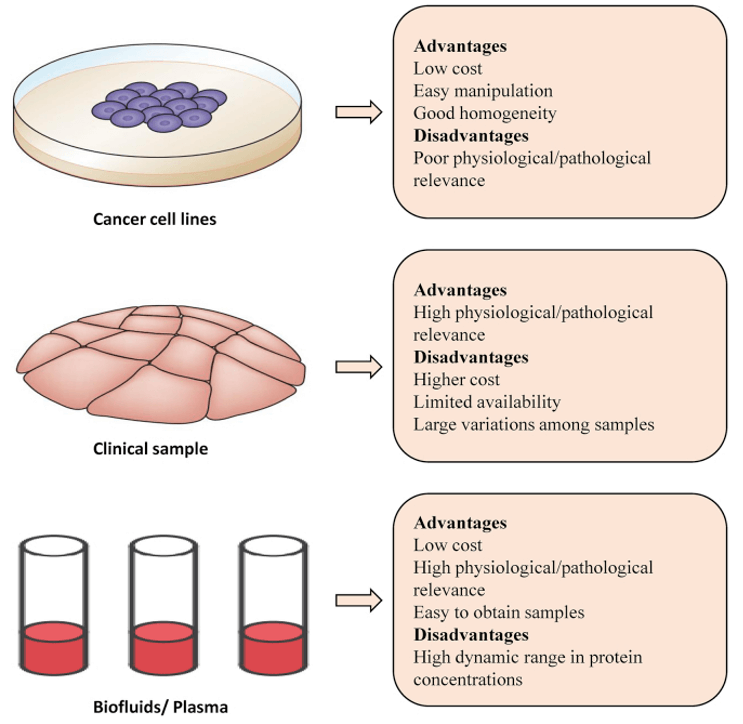 A comparison of the biological samples used in quantitative proteomics
A comparison of the biological samples used in quantitative proteomics
Currently, there are four major quantitative proteomics methods, namely labeled, label-free, targeted, and PTM methods, which have been widely used in cancer research. Among them, label-free quantification is simple to operate without labeling treatment, but requires high stability and reproducibility of experimental operations and it is suitable for large-scale quantitative comparisons and experimental designs where labeling quantification is not possible.
In recent years, mass spectrometry based on data-independent acquisition (DIA) has become an alternative technique for proteomics analysis of biological samples to reduce the limitations of data-dependent acquisition (DDA) analysis. In DDA-MS, the randomness of the automatic selection of precursor ions in the scan before fragmentation makes this method unable to repeatedly identify the same set of proteins in technical replication experiments. DIA is an unbiased method that allows peptide precursor ions to be split into several consecutive windows during fragmentation, thereby acquiring fragment ions of all precursor ions for accurate quantification of a given sample. Other benefits of DIA-MS based technology include reducing the number of clinical samples and providing sufficient proteomic coverage with quantitative consistency and analytical accuracy.
Applications of DIA-MS in Cancer Proteomics
Since DIA-MS was first published in 2012, the application of it in cancer proteomics has been steadily increasing. In 2019 alone, DIA-MS was used in 42 studies published in a series of cancer types to analyze various types of biological materials.
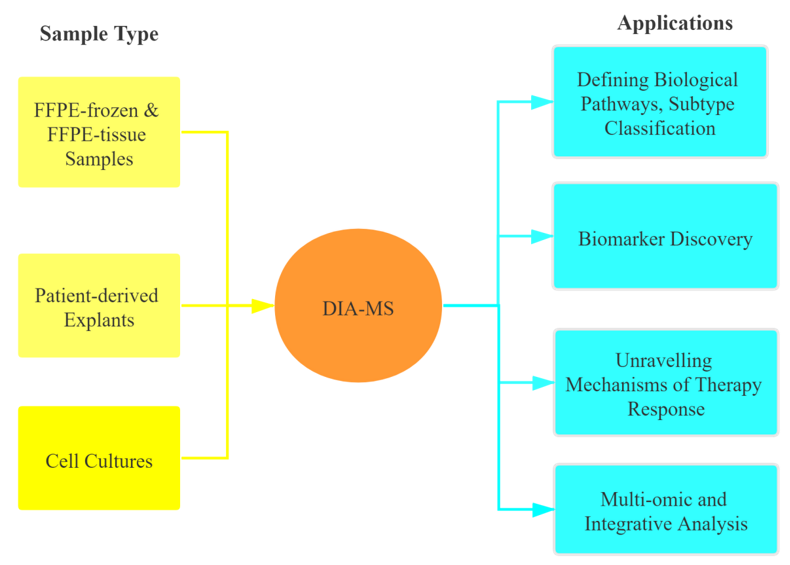
Our Cancer Proteomics Service
After a long period of dedicated research and development, Creative Proteomics has launched unique discovery proteomics and 4D-DIA services (DIA MS-based) to analyze the cancer proteome, enabling accurate and consistent quantification of DIA proteomics data to aid in the study of cancer research. Our Services can quantify up to 9,000 proteins in a single sample with high repeatability and accuracy, and are suitable especially for large cohorts of clinical samples. We also offer highly multiplexed targeted proteomics with absolute quantification for customized panels of proteins.
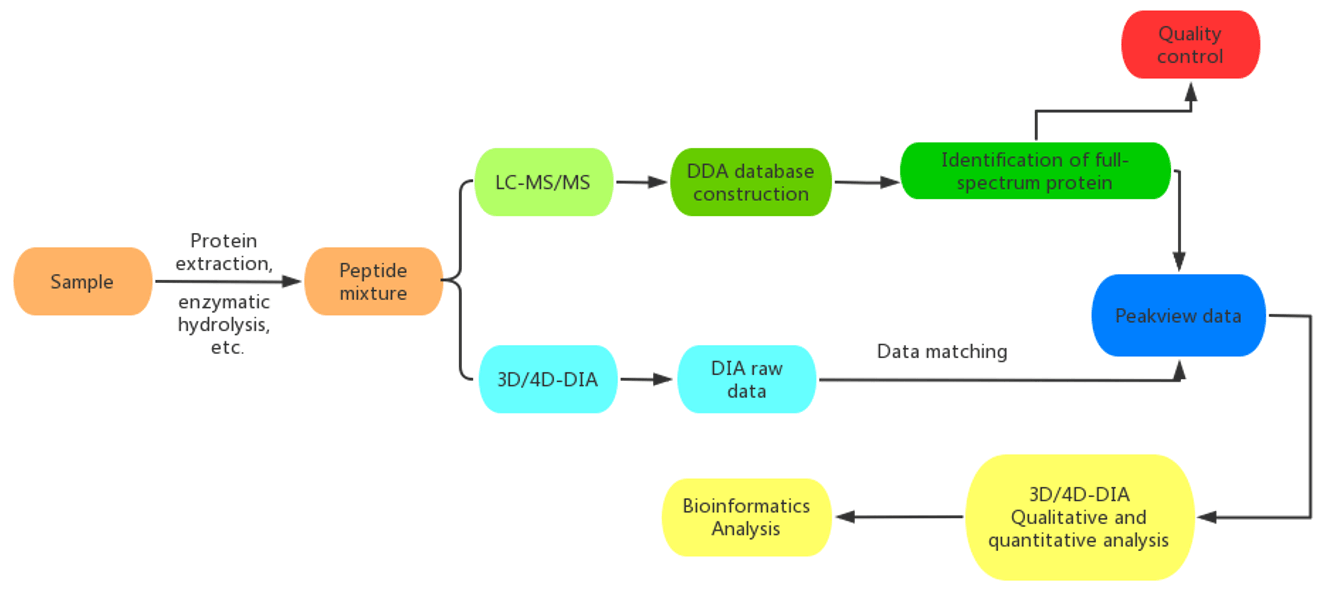
Workflow of Our Cancer Proteomics Service
Advantages
- High accuracy
- High throughput, up to 9,000 proteins can be identified at once
- Quantitatively identify nearly all detectable molecules, covering low-abundance proteins/peptides
- High repetition rate
- Complete and comprehensive information storage of samples in the first analysis
Sample Requirements
| Sample Type | Protein | # of Cell | Animal Tissue | FFPE | Blood | Urine | Serum | Microbes |
| Quantify | 100 ug | 1×107 cells | 1 g | 1mg fresh frozen
2 x 5um FFPE slices
Each 50-100nm2 | 1 mL | 2 mL | 0.2-0.5 mL | Dry weighed: 200 mg |
Report
- Experimental steps
- Relevant experimental parameters
- Mass spectrometry pictures
- Raw data
- Proteomics analysis results
Since we started to provide DIA technology service, we have accumulated a lot of experience not only in the detection of large cohorts of conventional samples, but also in providing one-stop service of DIA+PRM+machine learning for biomarker studies, tumor-based proteomic typing, and large-scale clinical sample proteomics analysis based on next-generation chromatography (Evosep DIA). Free consultation is welcome.
References:
- Lukas Krasny, Paul H. Huang. Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology. Molecular Omics, Issue 1, (2021)
- Yangying Zhou, et al. Proteomic signatures of 16 major types of human cancer reveal universal and cancer-type-specific proteins for the identification of potential therapeutic targets. Journal of Hematology & Oncology. 13, 170 (2020)
- Xiao-LiYang, et al. Quantitative proteomics characterization of cancer biomarkers and treatment. Mol Ther Oncolytics. 21: 255–263. (2021)
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 A comparison of the biological samples used in quantitative proteomics
A comparison of the biological samples used in quantitative proteomics


 4D Proteomics with Data-Independent Acquisition (DIA)
4D Proteomics with Data-Independent Acquisition (DIA)