Venom Protein Interaction Analysis
Venom proteins control all biological systems in a cell, and while many venom proteins perform their functions independently, the vast majority of proteins interact with others for proper biological activity. Creative Proteomics provides different venom protein interaction analysis services, characterizing protein–protein interactions through methods such as pull-down assays, co-immunoprecipitation (co-IP), crosslinking, far–western blot, and label transfer analysis is critical to understand venom protein function and the biology of the cell.
Introduction to Protein Interaction Analysis
Proteins in organism are the workhorses that facilitate most biological processes in a cell, including cell growth, gene expression, morphology, proliferation, nutrient uptake, apoptosis, intercellular communication and motility. And cells respond to a myriad of stimuli, and therefore protein expression is a dynamic process; the proteins that are used to complete specific tasks may not always be expressed or activated. In addition, all cells are not equal, and many proteins are expressed in a cell type–dependent manner. Protein-protein interaction refers to a process in which two or more protein molecules combine to form a protein complex. Under normal circumstances, it is difficult for a protein to function alone, ecause most protein molecules need the functions of each other to achieve complex functions. Therefore, focusing on the research of venom protein interaction is more conducive to exploring the pathogenic mechanism of venom, finding specific treatment methods for poisoning and providing new ideas in the research of new drug development.
What Creative Proteomics Offers
- Co-immunoprecipitation (Co-IP)
- Protein-protein interactions identified by pull-down experiments and mass spectrometry
- Protein interaction analysis of SILAC combined with immunoprecipitation and mass spectrometry
1. Co-immunoprecipitation (Co-IP)
Co-immunoprecipitation (CO-IP) uses the specificity between antigen and antibody to detect the physiological interaction between proteins. First, in the experiment, in order to maximize the retention of protein interactions in the cells, a non-ionic denaturant is used to lyse the cells. When the cell is lysed, the protein interaction in the cell is preserved, and the corresponding antibody is added to it to bind the antibody to the specific protein in the cell. Then add protein A/G to form an antibody-protein complex, perform centrifugation and separation to precipitate the antibody-protein complex, discard the supernatant, and finally use mass spectrometry to identify the precipitated protein to prove the interaction between the two. It is only suitable for analyzing stable or strong protein-protein interactions. It is usually used to identify the interaction between a target protein and another protein or to find unknown proteins that interact with a target protein.
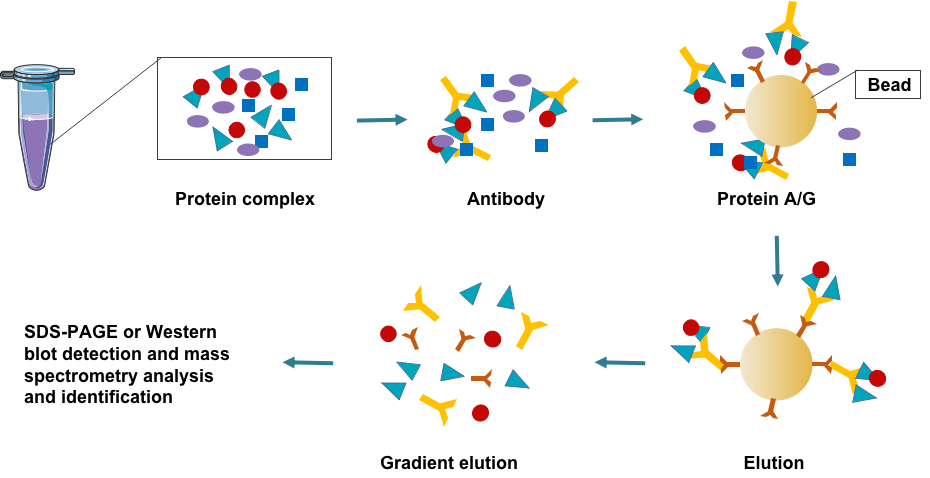 Fig 1. Creative Proteomics protein interaction analysis pipeline.
Fig 1. Creative Proteomics protein interaction analysis pipeline.
2. Protein-protein interactions identified by pull-down experiments and mass spectrometry
GST fusion protein pull-down sedimentation technology, using genetic recombination to insert a GST tag vector into the target protein A. Glutathione-coated magnetic beads bind to the target fusion protein, and after adding the cell lysate, the interacting protein is adsorbed by the fusion protein. Then add excess glutathione for elution, and finally analyze by MS technology. This method is mainly to study strong or stable protein-protein interactions in vitro. It can identify the possible direct interactions between two known proteins of interest; look for unknown proteins that may have an interaction relationship with the target protein.
3. Protein interaction analysis of SILAC combined with immunoprecipitation and mass spectrometry
The SILAC method was used to label the cells in the experimental group and the control group respectively, and then the Co-IP experiment was carried out. The immune complex was separated by the specific reaction between the antigen and the antibody. Then qualitatively/quantitatively detect the protein in the immune complex by LC-MS/MS. When the content of a certain protein in the experimental group and the control group reaches a statistical difference, it is determined that the protein has an interaction with the protein under study, which can greatly reduce the possibility of false positive results of protein interaction. The combination of SILAC technology and Co-IP-MS technology can be used for high-throughput quantitative analysis of protein interaction protein networks under specific conditions.
How to Order?
Thank you for your attention to our service! If you have any questions, please fill in the inquiry form and contact us immediately.
Reference
- Rao V S, et al. Protein-Protein Interaction Detection: Methods and Analysis[J]. International Journal of Proteomics, 2014, (2014-2-17), 2014, 2014:147648.
For research use only. Not intended for any clinical use.


 Fig 1. Creative Proteomics protein interaction analysis pipeline.
Fig 1. Creative Proteomics protein interaction analysis pipeline.