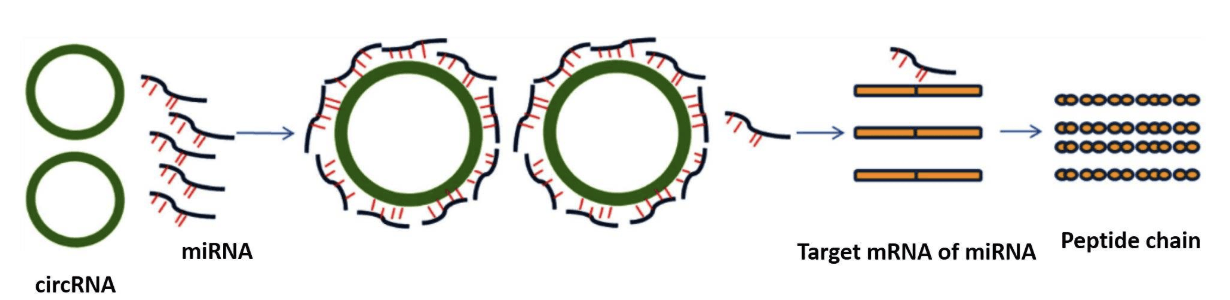
MiRNA is a class of evolutionarily conserved non-coding small RNA, with the function of regulating gene expression at the translation level. Mainly through the role of multiple target genes in many signal pathways, such as breeding, differentiation, proliferation, apoptosis. MiRNA can be combined with mRNA, to degrade the precepts and inhibit its translation and expression. In recent years, studies have also shown that lncRNA and circular RNA play the role of miRNA sponge in different species, which is called competitive endogenous RNA (ceRNA), can competitively combine with miRNA, to regulate the expression of target genes. The negative regulation association analysis of miRNA-transfer group, that is, according to the list of predicted interaction pairs of miRNA untargeted transplants, the results of miRNA differential screening and the differential transcript of specific modules (mRNA/lncRNA/circRNA), the negative regulation pairs among miRNA unspecific transducers were obtained, and the negative regulation network map of microRNA was constructed by cyctoscape.
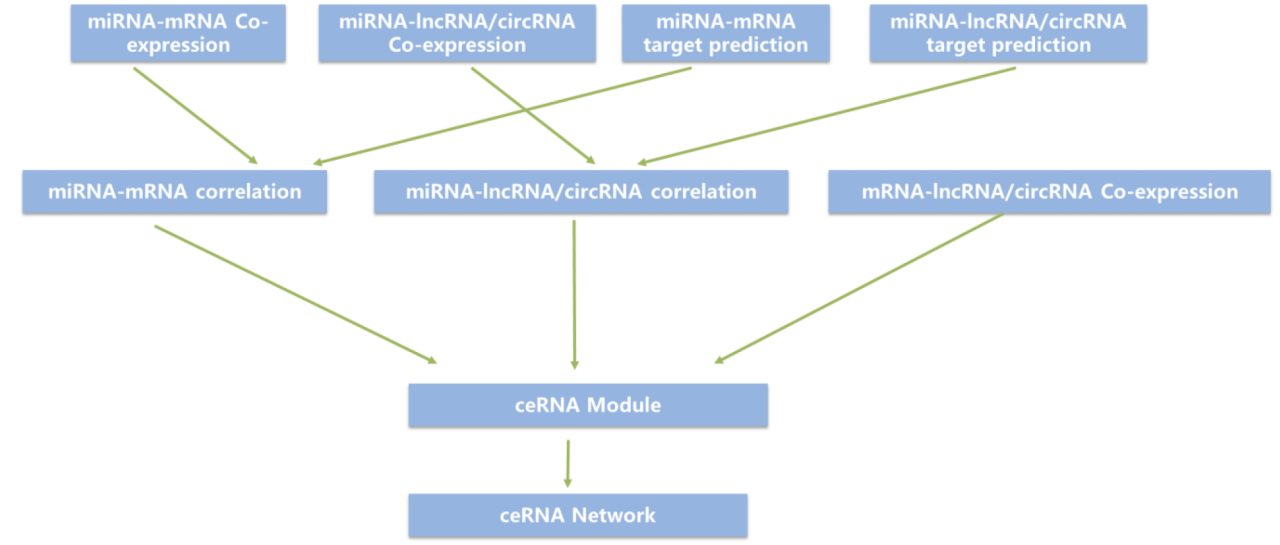 Figure 1. Technical route
Figure 1. Technical route
Step
[1] Prediction of target relationship between miRNA and transcripts (mRNA, lncRNA or circRNA): the samples were predicted by miranda software.
[2] Statistical test was used to obtain differential miRNA and differential transcripts (mRNA, lncRNA or circRNA), selected transcripts of special function modules).
[3] according to the differential screening file and the target relationship prediction file, the miRNA- transcripts with negative regulation relationship were obtained.
[4] according to the relationship pairs of miRNA- transcripts (mRNA, lncRNA or circRNA) with negative regulation, the network diagram was drawn by cytoscape software.
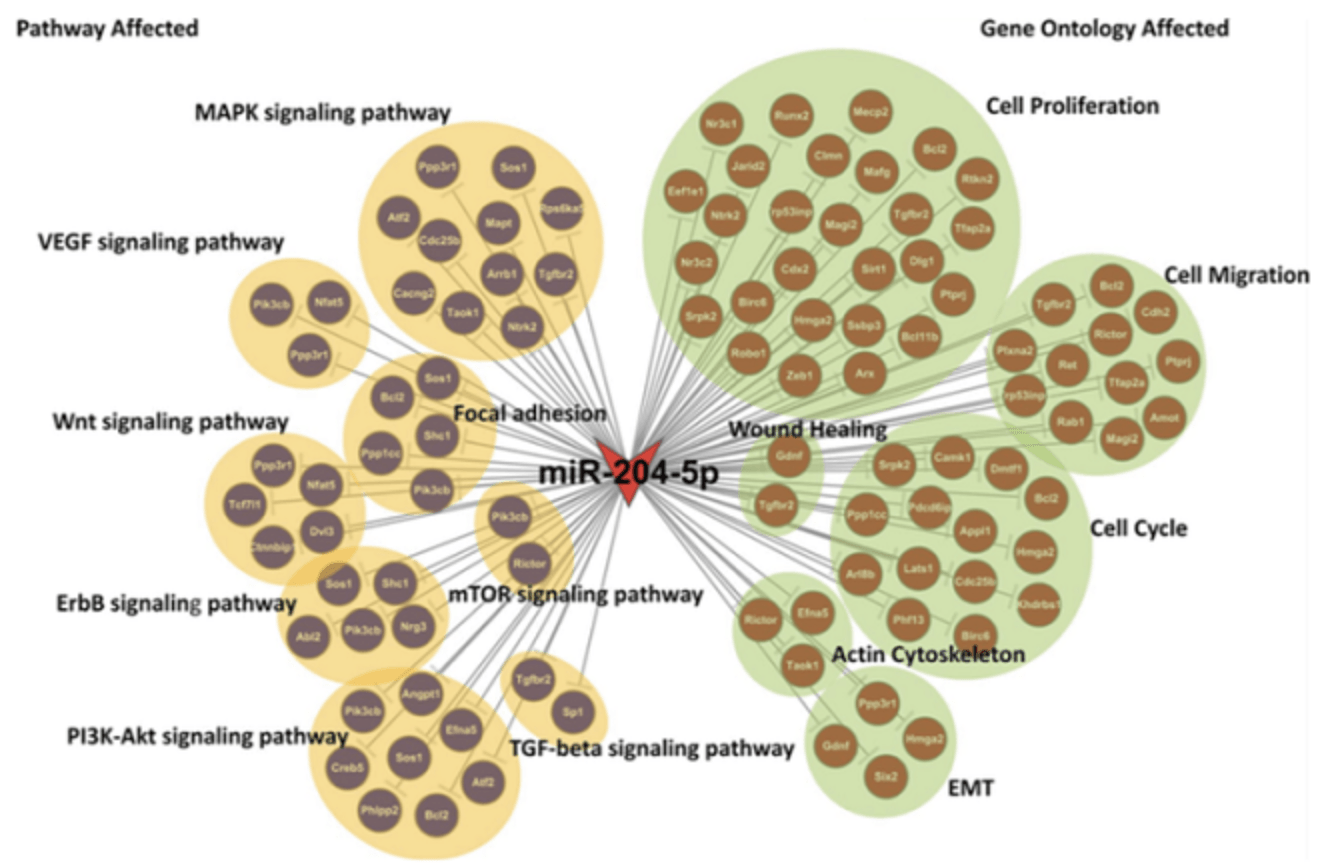
Figure 2. MiRNA target gene function and pathway network map (An J, et al. Investigative Ophthalmology & Visual Science, 2015)
The figure shows the genes targeted by miRNA-204, and the GO entries (right) and Pathway entries (left) that are significantly enriched by the target genes.
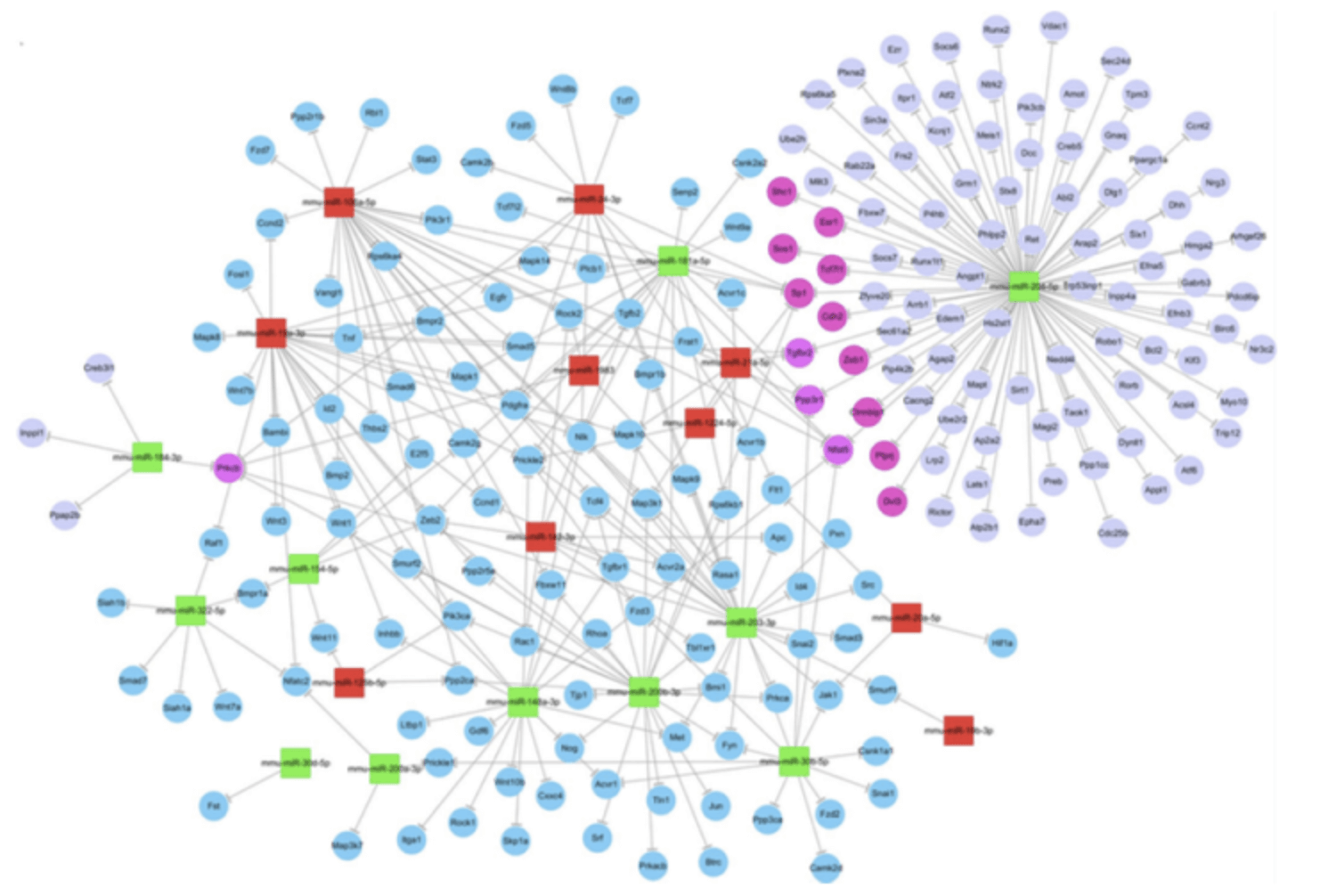
Figure 3. MiRNAs target gene network map (An J, et al. Investigative Ophthalmology & Visual Science, 2015).
Green indicates down-regulated miRNAs, red indicates up-regulated miRNAs.
Sample requirement.
Tissue samples: 1. Animal tissue: > 200mg; 2. Cell culture: > 2x106; 3. Whole blood: > 3ml;
RNA sample: 1. Please provide RNA; with a volume of 15 μL-100 μL, a total amount of ≥ 10 μg and a concentration of ≥ 200ng/ μL. 2. OD260/280 is between 1.8 and 2.2, OD260/ 230 ≥ 2.0, RIN ≥ 6.5, 28Spart 18s ≥ 1.0, to ensure that there is no degradation of RNA;
Our advantage
- A team of experts with decades of experience
- Powerful Bioinformatics Analysis Technology
- Joint analysis, mutual confirmation, enhance the persuasive power of the results
- To provide a comprehensive basis for the realization of accurate medical treatment.
- High quality service, high level of experiment, reliable analysis
Creative Proteomics, Negative Regulatory Association of miRNA-Transcriptomics "comprehensive attack", but also a full-length transcriptome sequencing necessary products, a full range of transcriptome sequencing services, for you to solve a variety of biological problems, to meet your diversified scientific research needs!
* For Research Use Only. Not for use in the treatment or diagnosis of disease.
Related Services:


 Figure 1. Technical route
Figure 1. Technical route