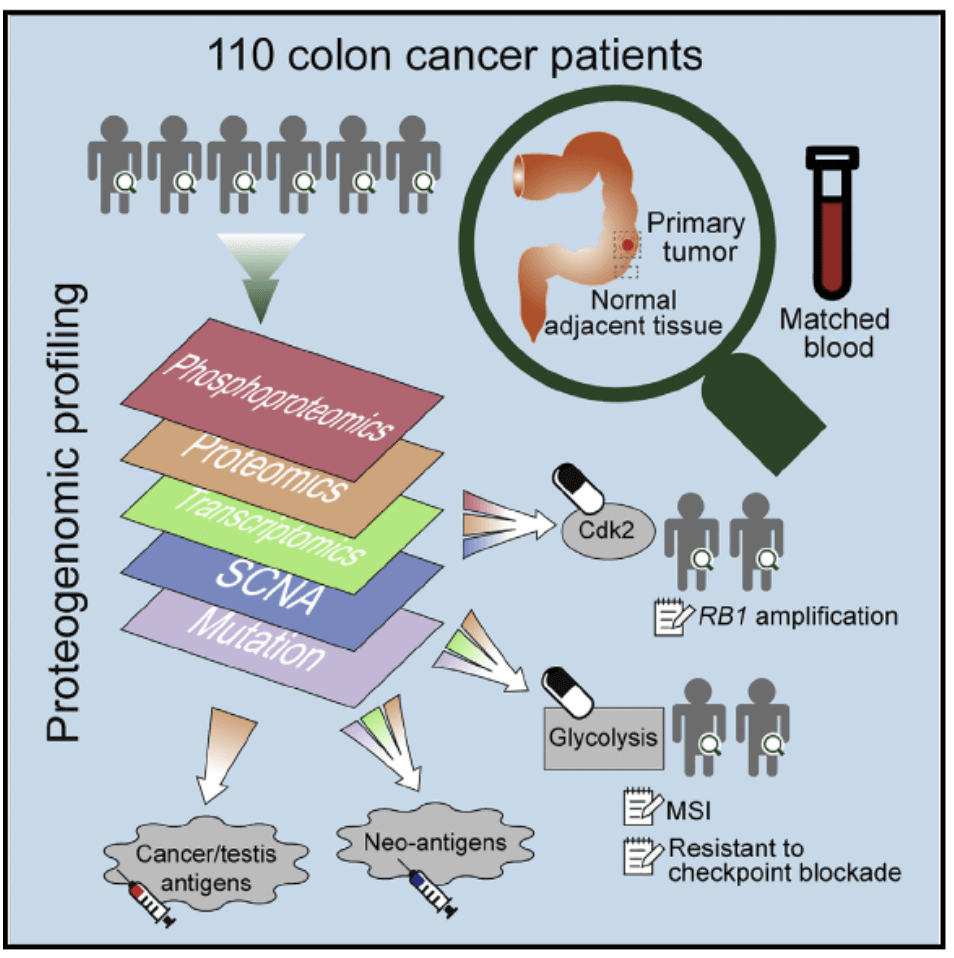
 Figure 1. Experimental flowchart(Vasaikar S, et al. 2019)
Figure 1. Experimental flowchart(Vasaikar S, et al. 2019)
In a new study, researchers have gained a more comprehensive understanding of the tumor by analyzing all the genes and proteins produced by colon cancer tissue from patients, and point out new cancer biological mechanisms and possible new treatment strategies. This study strongly supports a comprehensive characterization of tumor tissue as a means to guide further research in order to develop early diagnostic strategies and new treatments. The results are published in the May 2, 2019 issue of the journal Cell. The paper is titled "Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities."
Figure 2. Somatic Mutations and Their Proteomic Consequences(A and B) Significantly mutated genes in non-hypermutated (A) and hypermutated (B) samples.(Vasaikar S, et al. 2019)
The researchers first analyzed the newly discovered important mutant genes in MSI-H tumors and proteomics through somatic mutation analysis. The data shows unexpected functional complexity that cannot be predicted by mutation data alone. The truncated mutation removes the K398 ubiquitination site to stabilize the SOX9 protein and increase protein abundance. Functional assays support the carcinogenic effect of SOX9 in primary cells rather than its anti-cancer effect.In addition, the researchers learned that genomics and proteomics data complement each other, allowing scientists to better understand what is going on inside colon cancer cells.Researchers said, "An example is SOX9. Our genomic data set indicates that SOX9 is a tumor suppressor gene because it is frequently mutated in colon cancer in a way that indicates that the function of the protein encoded by this gene is disrupted and This protein is not produced or produced less. However, when we look at the proteomics data-the real protein in cancer tissues-we observe the opposite result; SOX9 protein is very abundant in these tumors That's more than normal. Proteomics data therefore challenge the notion that SOX9 is a tumor suppressor gene. "Further bioinformatics analysis provides possible explanations for this and other apparent contradictions between genomic and proteomic data, as well as other findings.
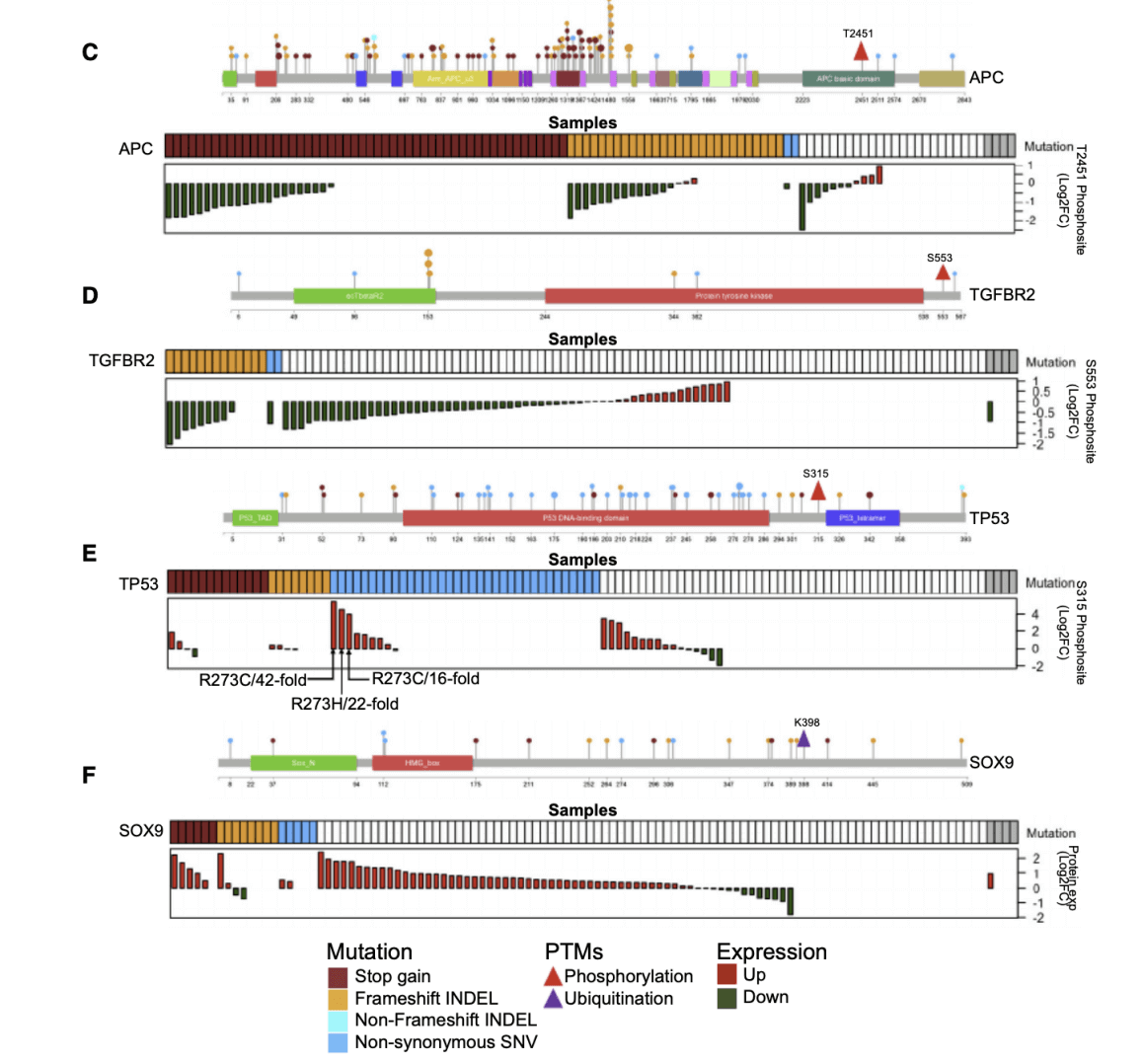 Figure 3. Somatic Mutations and Their Proteomic Consequences(A and B) Significantly mutated genes in non-hypermutated (A) and hypermutated (B) samples.
Figure 3. Somatic Mutations and Their Proteomic Consequences(A and B) Significantly mutated genes in non-hypermutated (A) and hypermutated (B) samples.
Second, the study used exome sequencing, microarray, RNA sequencing, microRNA sequencing, and proteomics techniques to prospectively assess mutations in a group of tissue samples (including tumor tissue and adjacent normal tissue) from patients with colon cancer , Copy number, gene expression, miRNA profiling, protein expression, and phosphoproteome levels to find new drug targets and potential tumor markers.By analyzing the characteristics of colon cancer subtypes, scientists revealed the potential role of glycolysis pathways in the resistance of immune checkpoint inhibitors, and provided new research ideas for our research on colon cancer. In addition, the colon cancer-associated protein-encoding genes found in the study have little overlap with known cancer genes, helping to develop personalized vaccines and other treatments.In addition to SOX9, there are other interesting new discoveries that may guide research to improve colon cancer treatment. For example, a biomarker called a DNA mismatch repair defect has been approved to identify candidate patients receiving immunotherapeutic therapy based on immune checkpoint inhibitors. However, only a portion of colon cancer patients with this biomarker respond to this therapy. Through bioinformatics analysis, new clues have been found as to why immunotherapy is not suitable for all colon cancers with defects in DNA mismatch repair, which may lead to the development of new treatments. The researchers provided all the data for people to access in a network tool called LinkedOmics. LinkedOmics also provides computing tools for further exploration of such data sets.
The researchers performed a proteomics study on the collected colon cancer cohort. Paired Proteome and Phosphorylated Proteomics Analysis of paired tumors and normal adjacent tissues has produced a catalog of proteins and phosphorylation sites associated with colon cancer, including known and putative new biomarkers, drug targets and cancer / Testicular antigen. Proteome integration can not only give priority to genomic inferred targets, such as copy number drivers and mutation-derived antigens, but also lead to new discoveries. Phosphorylation proteomics data correlates Rb phosphorylation with increased proliferation and decreased apoptosis in colon cancer. Proteomics identified association between decreased CD8T cell infiltration and increased glycolysis in microsatellite instability (MSI-H) tumors, suggesting that glycolysis overcomes MSI-H tumor resistance immune checkpoint block Potential targets. Proteomics provides a new way for biological discovery and therapeutic development.
Creative Proteomics has an excellent team of experts to provide you with customized multi-group oncology-related experimental design and experimental solutions, so that you can experience the highest quality, the most advanced and the most satisfactory service.Help scientists in the field of cancer research to produce quick results and good results, so as to promote scientific and technological innovation. Creative proteomics, multi-layer assemblage customization service experts to help you with your scientific research!
Reference
1. Vasaikar S, Huang C, Wang X, et al. (2019). "Proteogenomic Analysis of Human Colon Cancer Reveals New Therapeutic Opportunities." Cell. 177(4): 1035-1049.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 Figure 1. Experimental flowchart(Vasaikar S, et al. 2019)
Figure 1. Experimental flowchart(Vasaikar S, et al. 2019)