- Service Details
- Demo
- Case Study
- FAQ
- Publications
Identification And Quantification Of Plant Metabolomics
Plant metabolomics is more and more used for understanding physiological processes like the cellular responses to various stimuli. For example, in response to sulfur deficiency, the metabolites will be readjusted. It is found that there is a strict relationship between sulfur assimilation, nitrogen, lipid, and purine metabolism and enhanced photorespiration. What's more, metabolomics has been used for studying cold stress response. Metabolic engineering of biochemical pathways for pharmaceuticals production, gene function discovery are other applications of plant metabolomics.
Besides contributing to the color, taste, aroma, and scent of fruits and flowers, plant metabolites are associated with many resistance and stress responses in plants. As the end products of cellular regulatory processes, the levels of metabolites are the final response of biological systems to genetic alteration or environmental stimuli. These metabolites are strictly regulated by intracellular physiological fluctuations. Therefore, it is necessary to simultaneously identify and quantify metabolites in plants to understand the dynamics of the metabolites, study fluxes in metabolic pathways and decipher the role of each metabolite in response to various stimuli.
Because of their highly dynamic in time and space and complexity of structures, it is a big challenge to separate and characterize up to 200,000 metabolites in plant kingdom of high diversity in biological sample matrices. The studies of the plant metabolomics is composed of analysis of a large number of chemical species with various physical properties, ranges from ionic inorganic compounds to biochemically derived hydrophilic compounds, organic and amino acids, and a series of hydrophobic lipid-related compounds. Although technology is moving forward, we are still lack of knowledge of the plant metabolome. Because of the immense diversity among plants, metabolomics has become a crucial analytical tool in the plant community. It is difficult for a single analytical technique to identify and quantify all the metabolites found in plants. Plant metabolomics has benefited from a large number of previous methodological approaches and bioanalytical knowledge for the characterization of many chemically diverse classes of metabolites. While generally applicable methods for metabolite extraction, fractionation and detection is implemented, it is still challenging to identify and quantify the metabolites.
Detection Methods Of Plant Metabolomics
The classical instrumentation useful for the identification and quantification of plant metabolomics includes MS, GC, HPLC, or HPLC-MS, GC-MS. The coupling of chromatographic methods such as GC or HPLC with MS can greatly increase coverage of metabolites, which will enhance the biological context by increasing the number of identified metabolites. As one of the widely used analytical techniques in plant metabolomics, GC-MS is utilized to qualitatively and quantitatively analyze a wide range of volatile and derivatized nonvolatile metabolites with high thermal stability. Compared to LC-MS and LC-NMR, GC-MS is of analytical reproducibility and lower costs. LC-MS has been increasingly used in these years for the untargeted large-scale analysis of plant metabolomics, the characterization of metabolites in response to stress and the identification of flavonoid glycoconjugates in roots. LC-MS is more labile compounds and for compounds hard to derivatize. Besides, because of its advantage of high resolution and high reliability, CE-MS is also used for profiling of amino acid in plant cell cultures, for profiling of metabolites in bacterial-infected orange leaves and for profiling of metabolites in Illicium anisatum. Other techniques used for plant metabolomics include FT-IR, NIR, and NMR.
What Is Plant Untargeted Metabolomics
Plant untargeted metabolomics analysis ,also known as metabolite profiling in plants, is the most classical and widely used approach in plant metabolomics.It focuses on studying small molecule metabolite profiling in plants, investigating the metabolic products of different species, genetic types, or ecological types of plants at various growth stages or before and after specific stimuli. Based on liquid chromatography-mass spectrometry (LC-MS), this unbiased small molecule metabolites as possible within plant samples, comparing and analyzing experimental and control groups. Differential metabolites are selected through statistical analysis, followed by metabolic pathway analysis to explore the relative relationship between metabolites and physiological or pathological changes.
Workflow of Plant Untargeted Metabolomics
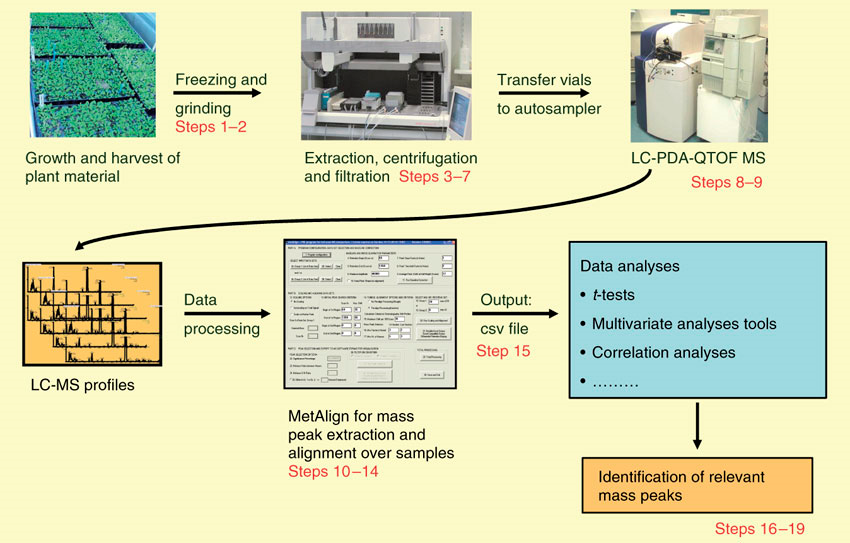
Applications of Plant Untargeted Metabolomics
- Plant Breeding Research
- Plant Growth and Development Mechanism Research
- Plant Stress/Abiotic Stress Mechanism Research
- Plant Variety Discrimination
- Evaluation of Agronomic Traits and Quality of Plants
- Construction of Secondary Metabolite Synthesis Pathways
- Plant Component Identification
- Evaluation of Agronomic Traits in Plants
- Study of Metabolite Accumulation Mechanisms in Different Plant Tissues
Our Plant Untargeted Metabolomics Service
With the integration of separation, characterization, identification, and quantification systems, which exhibit robustness, reproducibility, and high sensitivity, Creative Proteomics offers reliable, rapid, and cost-effective plant untargeted metabolomics using LC-MS. Simultaneously, both primary and secondary metabolites in plant samples are qualitatively and quantitatively analyzed, and plant metabolomic information is extensively explored using statistical and bioinformatics methods.
Our Plant Untargeted Metabolomics Service
Plant Primary Metabolism Network
Recent plant metabolomics research focuses on using primary metabolic networks to quantify yield and quality traits in crops like rice and maize. These traits are linked to complex primary metabolic pathways and are associated with quantitative trait loci (QTLs), offering insights for improving breeding accuracy.
Plant Secondary Metabolism Network
Metabolomics directly studies plant secondary metabolism by focusing on changes in compound content, typically the end products of metabolic pathways, without needing to account for factors like coenzymes or growth environments in the data analysis.
| Category | Quantity | Representative Substances |
|---|---|---|
| Amino Acids | 100+ | Serine, Threonine, Tyrosine, Valine, Glutamine, Cysteine, etc. |
| Vitamins | 30+ | Vitamin A, Vitamin B12, Vitamin C, Vitamin D2, Vitamin K1, etc. |
| Organic Acids | 200+ | Salicylic Acid, Malic Acid, Gallic Acid, Citric Acid, Tartaric Acid, etc. |
| Carbohydrates | 50+ | Lactose, Galactose, Sorbitol, Xylitol, Mannitol, Arabinose, etc. |
| Lipids | 100+ | Linoleic Acid, Palmitic Acid, Linolenic Acid, Myristic Acid, Arachidonic Acid, etc. |
| Nucleotides | 100+ | 1-Methyladenosine, 5-Methylcytidine, Adenine, Cytosine, Guanine, etc. |
| Phenols | 80+ | Danthron, Resveratrol, Emodin, Curcumin, Rhodiola Rosea Glycoside, etc. |
| Phenolic Acids | 60+ | Caffeic Acid, Catechin, Benzoyl Shanzhiside Methyl Ester, Danshensu, etc. |
| Flavonoids | 450+ | Baicalin, Isoflavones, Myricetin, Quercetin, Proanthocyanidin B1, etc. |
| Alkaloids | 370+ | Berberine, Galanthamine, Reserpine, Betaine, Ricinine, Barbaloin, etc. |
| Terpenes | 350+ | Astragaloside III, Ginsenoside, Betulin, Umbelliferone, Celosin, etc. |
| Quinones | 30+ | Danhong Danshensu I, Diosmetin, Chrysophanol, Danshensu IIA, Quercetin, etc. |
| Steroids | 60+ | Tomatidine, Dioscin, Digitalis Glycoside, Alliin, Levoalysine, etc. |
| Phenethylamines | 150+ | Verbascoside, Scopolamine, Caffeoylquinic Acid, 1-Caffeoylquinic Acid, etc. |
| Others | 400+ | Ganoderma Lucidum Triterpenoids, Podophyllotoxin, Rutin B, Acetylbenzene, Patchoulitones, etc. |
| Total | 2500+ |

Brochures
Metabolomics Services
We provide unbiased non-targeted metabolomics and precise targeted metabolomics services to unravel the secrets of biological processes.
Our untargeted approach identifies and screens for differential metabolites, which are confirmed by standard methods. Follow-up targeted metabolomics studies validate important findings and support biomarker development.
Download our brochure to learn more about our solutions.
Data Analysis Contents
| Basic Data Analysis | Advanced Data Analysis |
|---|---|
| Data preprocessing PCA analysis PLS-DA analysis OPLS-DA analysis Differential compound screening Differential compound identification | Metabolic pathway analysis Metabolic network analysis Multi-omics data correlation analysis (for studies with multiple omics data) |
Sample Submission Requirements
| Sample Type | Sample Size | Biological Replicates |
|---|---|---|
| Plant roots / stems / leaves / flowers / fruits / seeds / callus tissue / large fungi / root exudate freeze-dried powder | ≥ 200mg | 3~6 |
| Root exudate liquid | ≥ 500uL |

PCA plot

OPLS-DA plot

Heatmap plot

Volcano plot

Bar plot

KEGG analysis
Title: Quality evaluation of four Ferula plants and identification of their key volatiles based on non-targeted metabolomics
Journal: Frontiers in Plant Science
Published: 2024
Background
The study focuses on Ferula, a genus in the Apiaceae family, which is renowned for its medicinal and aromatic properties. Many species of Ferula are used for their pharmacological benefits, such as antibacterial, anti-inflammatory, and antioxidant effects. However, it is still unclear whether there is a distinction between edible and non-edible species of Ferula, which is crucial for the conservation and sustainable development of its resources.This study aims to systematically analyze the secondary metabolites in the leaves and roots of four Ferula species using untargeted metabolomics, in order to assess their edibility and medicinal value, and to explore the relationship between their unique flavor and medicinal components.
Materials & Methods
Sample preparation and measurement for untargeted metabolomics of roots
Samples were first prepared by extracting with methanol, vortexing, grinding, and ultrasound treatment. After centrifugation, the supernatant was filtered for analysis. UHPLC-MS/MS was used for metabolic profiling, with separation conducted on a UPLC column and detection via a Thermo Orbitrap Exploris mass spectrometer. The analysis was performed in both positive and negative ionization modes with gradient elution.
Metabolome data preprocessing and multivariate statistical analysis
For UHPLC-MS/MS data analysis, the data was converted to mzXML format and processed using the MSConvert tool. The R XCMS software package was used for peak detection, filtering, alignment, and data analysis. Identification of substances was carried out by referencing public databases like HMDB, MassBank, LipidMaps, mzCloud, KEGG, and a self-built material bank, with a parameter of ppm < 30. Data were corrected using LOESS signal correction and normalized, with only ion peaks with relative standard deviations (RSDs) less than 30% retained. MetaboAnalyst was used for principal component analysis (PCA), orthogonal partial least squares discriminant analysis (OPLS-DA), and clustering heatmaps to analyze data. Differential metabolites were screened based on Fold Change ≥ 2 or Fold Change ≤ -2 and p < 0.05, with further analysis using VIP > 1.0 for root metabolites. Finally, variance analysis (ANOVA), significance testing (p < 0.05), and mapping were conducted using SPSS 19.0 and Origin 2023b software.
Results
Analysis of the metabolic products in the roots of four Ferula plants.
The authors used an untargeted metabolomics approach to analyze root samples from four edible and non-edible Ferula species, identifying a total of 17 classes and 709 metabolites. Hierarchical clustering analysis of the results revealed significant overall differences between the roots of different Ferula species. To further validate the differences between the edible and non-edible groups, multivariate analysis was performed on the metabolite data using MetaboAnalyst. PCA showed that the samples from the two groups could be clearly distinguished, with the sum of the contributions from the first two principal components in both positive and negative ion models exceeding 70%, indicating that the model was reliable. This was consistent with the heatmap results.
 Figure 7.Metabolite analysis of Ferula root, showing the distribution of metabolites, clustering patterns, and multivariate analysis results.
Figure 7.Metabolite analysis of Ferula root, showing the distribution of metabolites, clustering patterns, and multivariate analysis results.
Metabolic pathway analysis and interaction study
The authors used MetaboAnalyst to perform pathway enrichment analysis on differential metabolites, revealing that 42 volatile compounds and 62 untargeted metabolites were enriched in 30 metabolic pathways, with key pathways including phenylalanine metabolism and fatty acid biosynthesis. Both pathways were significant for flavor and medicinal value in Ferula. A correlation analysis showed that volatile metabolites from Ferula feurlaeoides negatively correlated with amino acid metabolites, while those from other edible species positively correlated, suggesting amino acid metabolism plays a central role in the synthesis of Ferula's aromatic compounds.
 Figure 8. KEGG pathway classification of volatile and root metabolites, and correlation analysis of key metabolites between the roots and volatiles of edible and non-edible Ferula.
Figure 8. KEGG pathway classification of volatile and root metabolites, and correlation analysis of key metabolites between the roots and volatiles of edible and non-edible Ferula.
Conclusion
This study systematically analyzed the metabolites in the leaves and roots of four Ferula species using untargeted metabolomics. The results identified 166 volatile metabolites in the leaves and 1,079 metabolites in the roots. Additionally, 42 potential volatile metabolites and 62 differential root metabolites were selected to distinguish edible and non-edible Ferula species. The findings reveal the relationship between Ferula's unique flavor and medicinal components, identifying sulfur, aldehyde, and ketone-containing compounds as the primary contributors to its strong odor. These results provide a theoretical basis for selecting edible Ferula species and assist in evaluating the quality of different Ferula species.
Reference
- Jiang, M., Peng, M.,et.al. (2024). Quality evaluation of four Ferula plants and identification of their key volatiles based on non-targeted metabolomics. Frontiers in plant science, 14, 1297449. https://doi.org/10.3389/fpls.2023.1297449
What is metabolic profiling of plants?
Metabolic profiling of plants involves the systematic analysis of the metabolites produced by a plant. This process identifies and quantifies the small molecules (metabolites) such as sugars, amino acids, lipids, and secondary metabolites that are involved in plant metabolism. By examining these metabolites, researchers can gain insights into plant health, growth conditions, genetic makeup, and stress responses, helping with crop improvement, medicinal plant research, and sustainable agriculture.
What are plant primary metabolites and secondary metabolites?
Primary metabolites are essential compounds involved in basic cellular functions, such as growth, energy production, and reproduction. These include amino acids, fatty acids, carbohydrates, and nucleotides.Secondary metabolites are compounds produced by plants that are not directly involved in growth or reproduction but play key roles in defense, communication, and attraction. These include alkaloids, flavonoids, terpenoids, and phenolics. Secondary metabolites often have medicinal, aromatic, and toxic properties.
What is the HPLC method for plant extracts?
High-Performance Liquid Chromatography (HPLC) is a widely used analytical technique for separating, identifying, and quantifying components in plant extracts. It works by passing a liquid sample through a column filled with a stationary phase. The different components in the extract interact differently with the stationary phase and are separated as they pass through. This method is particularly useful for analyzing complex mixtures of plant compounds like alkaloids, flavonoids, and essential oils.
What metabolic pathways do plants use?
Plants utilize various metabolic pathways to sustain life, produce energy, and synthesize essential compounds. Key pathways include photosynthesis, which converts light energy into chemical energy, and glycolysis and the citric acid cycle (Krebs cycle), which generate energy from carbohydrates, fats, and proteins. Additionally, plants have secondary metabolic pathways that produce bioactive compounds like flavonoids, terpenoids, and alkaloids, which play roles in defense, growth regulation, and attracting pollinators. These metabolic networks allow plants to adapt to environmental conditions and ensure their survival.
Water-soluble saponins accumulate in drought-stressed switchgrass and may inhibit yeast growth during bioethanol production
Chipkar, S., Smith, K., et al
Biotechnology for Biofuels and Bioproducts
Year:2022








