- Service Details
- Demo Results
- Case Study
What Is Disulfide Bond
Disulfide bonds are chemical bonds that connect different peptide chains or different cysteine residues within the same peptide chain. They are a post-translational modification in proteins, formed between the sulfur atoms of two cysteine residues during the protein's biosynthesis process within cells. Disulfide bonds are relatively stable covalent bonds that play a role in stabilizing the spatial structure of peptide chains within protein molecules. The greater the number of disulfide bonds, the greater the stability of the protein molecule against external influences.
Disulfide bonds (S-S bonds) are formed through the oxidation of sulfhydryl groups (-SH) on two cysteine residues within a protein. They are an important form of post-translational modification in proteins. Proteins are interconnected by various interchain and intrachain cysteine linkages, which are essential for maintaining the correct tertiary structure of protein molecules and preserving their biological activity. The arrangement of disulfide bonds in antibody drugs reflects their structural characteristics. Improper formation or interchange of disulfide bonds can lead to antibody aggregation, highlighting the crucial role of confirming the pattern of disulfide bond connections in the process of confirming the structure of antibody drugs. Modern mass spectrometry techniques have become important tools for disulfide bond analysis due to their simplicity, speed of analysis, and high sensitivity.
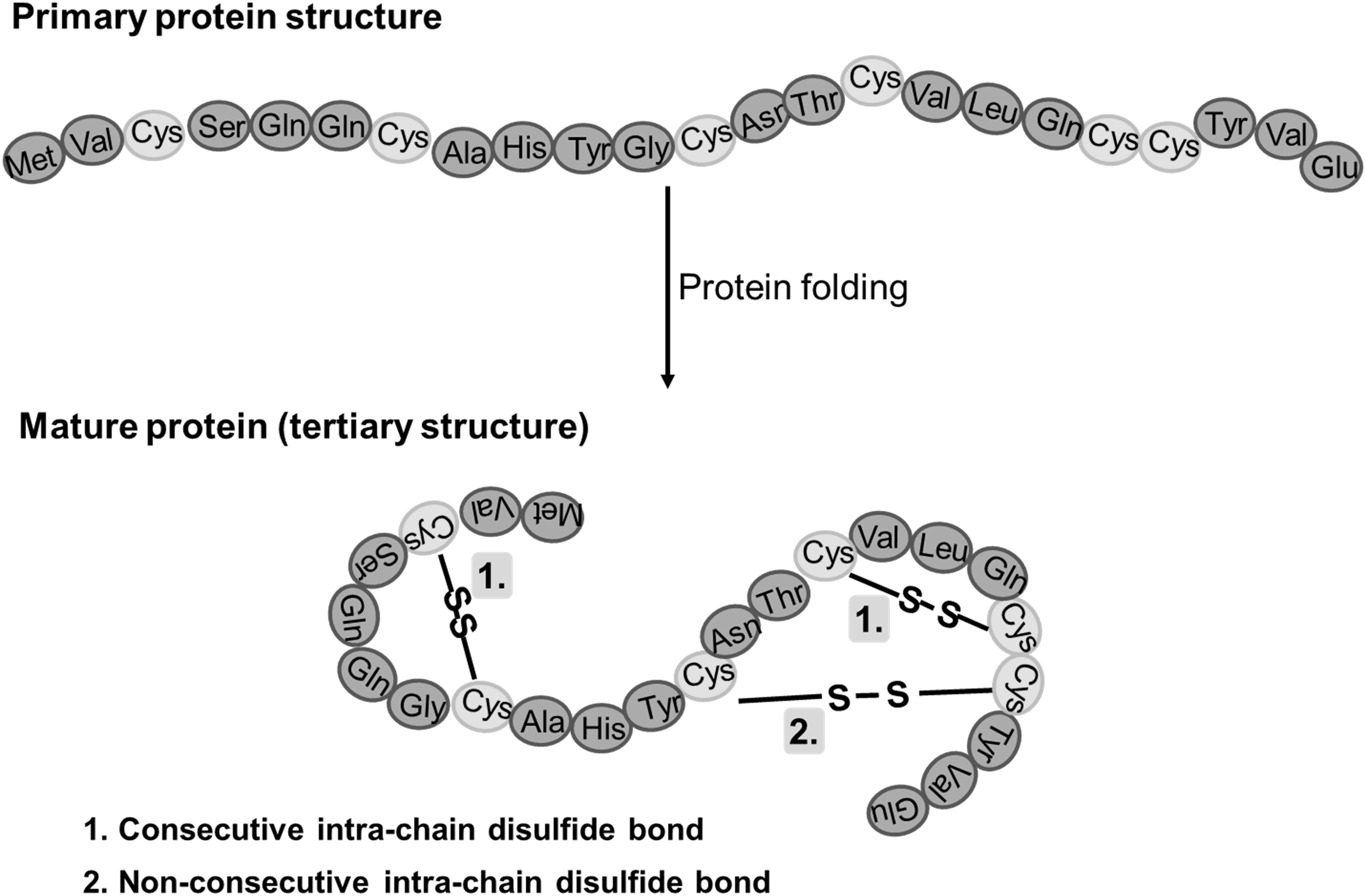
Figure 1. disulfide bonds in mature proteins
Disulfide Bond Analysis Process
Creative Proteomics provides services to map the disulfide bond sites of unknown proteins and verify the disulfide bonds and folding structure of protein samples. Basic steps include peptide extraction, peptide separation and data analysis.
- Peptides extraction: We have developed mature sample processing technology, such as reducing the chance of in vitro disulfide bond exchange and maintaining the native protein structure to the greatest extent.
- Peptides analysis: We can map disulfide bonds by utilizing LC-MS/MS. We have developed an advanced platform containing Orbitrap Fusion, Q Exactive HF, Orbitrap Fusion Lumos.
- Date analysis: Different software (like pLink-SS and SlinkS) is used to achieve disulfide bond connection and free sulfur analysis. In addition, bioinformatics analysis will also be performed.
- Detailed reports. We provide technical reports containing experiment procedures, parameters of liquid chromatography and mass spectrometer, MS raw data files, peptide identifications and intensities, protein identifications and disulfide bonds mapping and bioinformatics analysis.
Sample Requirement
A variety of biological samples are available for testing, such as cell, animal tissue, blood, serum and so on.
| Types | Volume |
|---|---|
| Protein | 100ug |
| Cell | 2×107 cells |
| Animal Tissue | 1g |
| Plant Tissue | 200mg |
| Blood (EDTA added) | 1ml |
| Serum | 0.2-0.5ml |
| Urine | 2ml |
| Microbes | Dry weighed: 200mg |
| If you want to know specific sample requirements, please feel free to contact us. | |
Advantages
- Available for protein analysis at the single protein level and proteomics level
- Highly versatile. We can analyze various forms of samples such as tissue extracts, whole cell lysates, subcellular fractions and so on
- High throughput, high degree of automation, and strong separation ability
Experimental Instrument
Q Exactive HFX Mass Spectrometer
Principle of Disulfide Bond Mass Spectrometry Analysis
Combining non-reducing enzymatic digestion within protein solutions and utilizing nanoLC-MS/MS detection, the analysis of disulfide bond pairing in the sample is conducted. This process clarifies the various intrachain and interchain disulfide bond pairings within the sample and provides empirical evidence for determining these disulfide bond pairings, including primary and secondary mass spectrometry data.

- Exemplifying Precision and Accuracy: Proficiently discerns the exact locations of disulfide bond peptide sites, effectively countering the potential for false positive outcomes.
- Versatility in Analysis: Demonstrates the ability to scrutinize diverse forms of disulfide bond configurations.
- Efficient and User-Intuitive Functionality: Employs a streamlined pre-processing approach, obviating the necessity for supplementary experimental steps.
Applications of disulfide bond analysis service:
- Study unknown disulfide bonds in novel proteins.
- Analyze disulfide bonds in refolded proteins to test whether a protein is correctly folded.
- Confirmation of correct disulfide bond linkage in protein therapeutics.
Technology platform
- Ion Chromatography
- High Performance Liquid Chromatography (HPLC)
- Matrix Assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS)
We are specialized in quantitative multiplexed proteomics and metabolomics applications through the establishment of state-of-the-art mass spectrometry platforms, and we have optimized sample processing methods and a series of advanced analytical methods that provide a strong guarantee for the success of the research. As every project has different requirements, please contact our specialists to discuss your specific needs. We are looking forward to cooperating with you.
The specimens under investigation comprise either protein or peptide samples, with emphasis on gauging the molecular weight of the peptide molecule encompassing disulfide bonds. The determination entails discerning both the amino acid sequence and the specific disulfide bond pairing arrangement within said peptide entity.

Streamlining the Characterization of Disulfide Bond Shuffling and Protein Degradation in IgG1 Biopharmaceuticals Under Native and Stressed Conditions
Journal : Front Bioeng Biotechnol
Published: 2022
Post-translational modifications (PTMs) have been shown to negatively impact the efficacy and safety of proteins by altering their native conformation, stability, target binding, and pharmacokinetics. The formation of disulfide bonds aids in proper protein folding and contributes significantly to normal protein function. Disulfide bond mispairing, however, can reduce drug stability, decrease efficacy, and increase immunogenicity. Therefore, regulatory bodies such as the FDA, EMA, and ICH designate disulfide bonds as critical quality attributes (CQAs) in biopharmaceuticals, with ICH also noting disulfide bond mispairing as a common mechanism of protein degradation.
This study investigates changes in disulfide bond mispairing ratios of monoclonal antibodies during stress stability using LC-MS/MS. The research combines results from comparative size exclusion chromatography (SEC) and SDS-PAGE to characterize the degradation trend of biopharmaceuticals. This research offers a standardized workflow for biopharmaceutical companies and regulatory agencies, providing direction for the identification and quantification of disulfide bonds and mispairing in biopharmaceuticals.
Experimental Protocol
The batches of samples, including Rituximab Originator (Rit OR), Rituximab Biosimilar (Rit BS), Bevacizumab Originator (Bev OR), and Bevacizumab Biosimilar (Bev BS), were divided into three portions each. One portion served as a control, while the other two portions were separately incubated at 37°C and 240rpm for 2 weeks and 4 weeks. After denaturation and NEM blocking at all time points, enzymatic digestion was performed using Trypsin Platinum and rLys-C under pH=5.4 conditions. LC-MS analysis, utilizing the Byos Disulfide Bond workflow, was conducted to assess and quantify disulfide bond mispairing. Verification was accomplished through complementary SEC and SDS-PAGE analyses.
 Figure 2: Disulfide bond analysis conducted using the Byos Disulfide Bond workflow.
Figure 2: Disulfide bond analysis conducted using the Byos Disulfide Bond workflow.
Experimental Results
LC-MS Findings: Initially, all four sample batches exhibited low levels of disulfide bond mispairing. Generally, both the originator and biosimilar forms of the two monoclonal antibodies displayed a gradual increase in disulfide bond mispairing under the aforementioned stress conditions over time. The specific observations are as follows:
Rituximab Originator: Disulfide bond mispairing increased from 0.24±0.21% (baseline) to 0.51±0.11% (4 weeks).
Rituximab Biosimilar: Disulfide bond mispairing increased from 0.27±0.07% (baseline) to 0.36±0.08% (4 weeks).
Bevacizumab Originator: Disulfide bond mispairing increased from 0.58±0.08% (baseline) to 1.46±1.10% (4 weeks).
Bevacizumab Biosimilar: Disulfide bond mispairing decreased from 1.62±0.78% (baseline) to 1.10±0.50% (2 weeks), then increased to 1.25±0.20% (4 weeks). The abnormal intermediate increase could possibly be attributed to factors such as small sample size, which may not align with the SEC and SDS-PAGE test results.
 Figure 3: Disulfide Bond Mispairing Results Compilation
Figure 3: Disulfide Bond Mispairing Results Compilation
SEC and SDS-PAGE Results: Under the aforementioned stress conditions, SEC analysis revealed a degradation trend over time in all four sample batches. However, the distinction lies in the observed formations – Rituximab resulted in fragmentation, whereas Bevacizumab formed aggregates. This trend was further substantiated by SDS-PAGE gel electrophoresis patterns.
 Figure 4: Representative SEC chromatograms
Figure 4: Representative SEC chromatograms
 Table 1: SEC data
Table 1: SEC data
 Figure 5: SDS-PAGE results
Figure 5: SDS-PAGE results
Conclusion and Discussion
Through the implementation of stress conditions at 37°C and 240rpm, the authors comprehensively assessed the alterations in two IgG1 monoclonal antibodies, both originator and biosimilar versions, using a tri-dimensional approach encompassing LC-MS/MS, SEC, and SDS-PAGE. The observed congruence between the trends in disulfide bond mispairing and degradation suggests a potential correlation, thereby offering insights into the identification, characterization, and quantification of protein disulfide bonds.
References
- Bošnjak I, Bojović V, Šegvić-Bubić T, et al. Occurrence of protein disulfide bonds in different domains of life: a comparison of proteins from the Protein Data Bank[J]. Protein engineering, design & selection, 2014, 27(3): 65-72.
- Coghlan J, Benet A, Kumaran P, Ford M, Veale L, Skilton SJ, Saveliev S, Schwendeman AA. Streamlining the Characterization of Disulfide Bond Shuffling and Protein Degradation in IgG1 Biopharmaceuticals Under Native and Stressed Conditions. Front Bioeng Biotechnol. 2022 Mar 14;10:862456. doi: 10.3389/fbioe.2022.862456. PMID: 35360407; PMCID: PMC8963993.















