As a post-translational modification of proteins, crotonylation plays an important role in chromatin remodeling, cell metabolism, cell cycle, and cellular reorganization. Histone crotonylation is a newly identified covalent epigenetic modification, which is a dynamic process analogous to histone acetylation regulated by histone crotonyl transferases (HCTs) and histone decrotonylases (HDCRs). Meanwhile, lysine crotonylation has been proved to have the function of promoting gene expression. After introducing the negatively charged crotonyl groups, positive charges of histones were neutralized and bound more loosely to negatively charged DNA. Therefore, this overall process ameliorates the binding of transcription factors. Nevertheless, histone crotonylation modifications have been shown to maintain the activity of sex chromosome-associated genes in mid-meiotic divisions.
Although initially identified on histones, lysine crotonylation has expanded to a great number of non‐histone proteins. In order to characterize the global crotonylation proteome, proteomic solutions based on sensitive immune‐affinity purification and high‐resolution liquid chromatography‐tandem (LC‐MS/MS) were applied to identify new crotonylated proteins and modification sites. In addition, the utilization of antibodies with high specificity targeting crotonylated peptides involved in immunoprecipitation significantly advanced the enrichment and identification of crotonylated lysine residues[1].
Our Crotonylation Proteomics Service
Based on our advanced mass spectrometry instrumentations and extensive knowledge in biochemical, Creative Proteomics has launched a crotonylation proteomics service package, which identifies crotonylated proteins and corresponding crotonylation sites for complex samples such as tissues and cells.
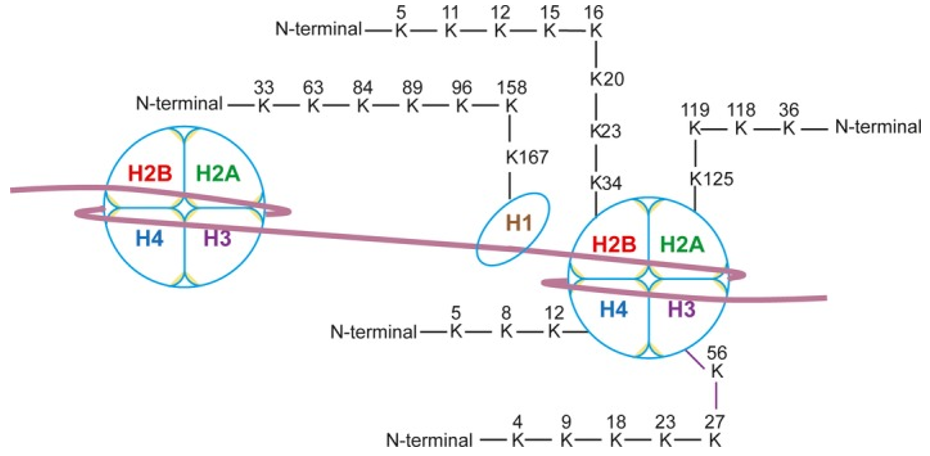
Fig. 1 Human histone crotonylation sites[1]
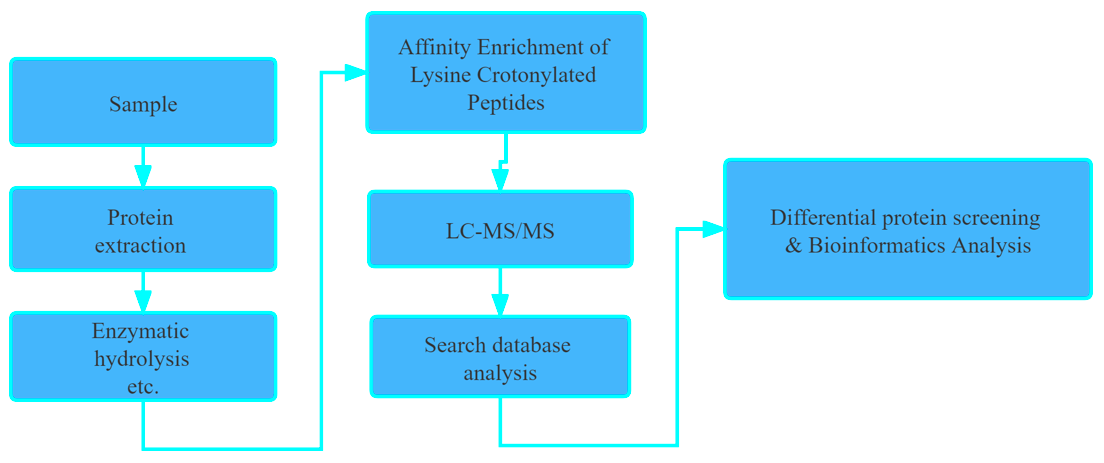
Workflow
Customized Bioinformatics Services
| Standard Analysis | Customized Analysis |
|---|---|
a) Collection of output statistics | Modified peptide motif analysis |
Sample requirements
- Fresh animal tissue: ≥600 mg
- Fresh plant tissue: ≥6 g
- Cell culture: ≥1×107 cells/tube x 3 tubes
- Fungi and bacteria: ≥600 mg
- Serum, plasma: 450 μl × 4 tubes
- Protein solution: total protein of 5-10 mg
- Body fluid samples: urine of 15 mL × 4 tubes (centrifuge at 1000 x g for 5 minutes and discard sediment); or other body fluids (saliva, amniotic fluid, cell culture supernatant, etc.) > 15 mL
Advantages
- High specificity and enrichment efficiency by using a chemically stable crotonylated resin IP
- Large-scale identification of enriched crotonylated peptides with mass spectrometry of high resolution and high throughput
- Combining commercially available quantitative techniques to analyze, compare, and correlate crotonylation levels between samples quantitatively
Applications
- Drug target research
- Disease biomarker screening
- Plant stress/resistance research
- Mechanism of action studies
Technology platforms
Ion Chromatography
High-Performance Liquid Chromatography (HPLC)
Matrix-Assisted Laser Desorption Ionization Mass Spectrometry (MALDI-MS)
References
- Junhu Wan, Hongyang Liu, Jie Chu, Hongquan Zhang. Functions and mechanisms of lysine crotonylation. J Cell Mol Med. 2019 Nov; 23(11): 7163–7169.
- Gaoyue Jiang, Chunxia Li, Meng Lu, Kefeng Lu & Huihui Li Protein lysine crotonylation: past, present, perspective. Cell Death & Disease 12, 703 (2021).
- Shuo Wang1, Guanqun Mu1, Bingquan Qiu1, Meng Wang1, Zunbo Yu2, Weibin Wang3, Jiadong Wang3, Yang Yang. The Function and related Diseases of Protein Crotonylation. Int J Biol Sci 2021; 17(13):3441-3455.







