Equipped with state-of-the-art technologies such as high-performance liquid chromatograph coupled to tandem mass spectrometry (LC-MS/MS) and advanced computational algorithms, Creative Proteomics can offer accurate and fast antibody de novo sequencing service customized to your needs.
De novo sequencing of antibodies refers to the analytical process of deriving the amino acid sequence of an antibody and discovering any associated post-translational modification without knowing DNA or protein sequence beforehand.
Our De Novo Antibody Sequencing Workflow
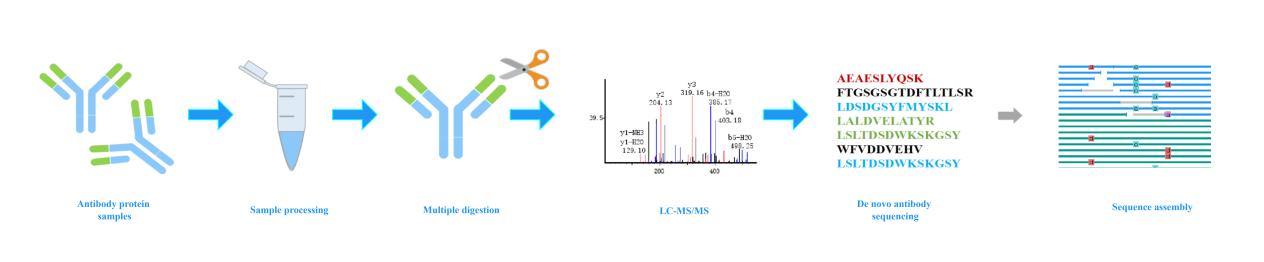
In a typical workflow, the antibody is digested by using a panel of different enzymes (either one at a time or a combination of a few) including the most commonly used one: trypsin and several others such as Glu-C, Asp-N, chymotrypsin and etc. If required, the heavy and light chains of the antibody can be separated prior to enzymatic digestion. Then the digested peptides are analyzed by LC-MS/MS. Creative Proteomics uses hybrid quadrupole-Orbitrap mass spectrometers for data acquisition, which generates data with superior quality: high resolution and accuracy. Lastly, our senior specialists perform data analysis by using the most advanced computational algorithms, which efficiently derive the amino acid sequence of digested peptides and assemble overlapped peptides. Eventually, a comprehensive result report including raw MS data is provided to our customers.
Our De Novo Antibody Sequencing Service
We only need a few hundred micrograms of an antibody with a minimum purity of 95% for de novo antibody sequencing. Leucine and isoleucine are assigned based on preferences of enzyme cleavage sites and frequencies in our database.
Besides the primary amino acid sequence of heavy and light chains of the antibody, Creative Proteomics is also able to characterize post-translational modifications such as glycosylation and others according to customers’ requests. Please contact us if you are interested in sequencing your antibody of interest and our experts are willing to help and discuss your needs in detail.
Advantages
Applicable to different species of antibodies and different antibody isoforms
Quantification of post-translational modifications and variant sequences
Our antibody sequencing platform can effectively distinguish between different variants of homologous amino acids, enabling accurate determination of leucine and isoleucine.
For post-transcriptional modification of amino acid, our system can also accurately distinguish.
Strict quality control
Our antibody sequencing technology can also accurately complete the sequencing of antibodies labeled with small groups, such as FITC, Biotin, etc., and can also complete the sequencing of antibodies labeled with large protein groups such as GST.
High accuracy
Our antibody sequencing platform makes it easier and more accurate to differentiate amino acids that are difficult to distinguish between molecular weights by traditional techniques, such as the analysis of glutamine and lysine with a molecular weight difference of 0.03 Da.
High antibody sequence coverage
We use up to 8 different proteases to digest and digest the target antibody for different project needs, and use PEAKS software to achieve the accuracy of full sequence splicing of digested peptide segments of different light and heavy chain.
For the CDR region, peptide sequences containing a single site are generated 10 times to ensure accurate subsequent antibody sequence splicing through high coverage.
Sequencing of fluorescently labeled monoclonal antibodies, Immunoglobulin M, and other non-traditional antibodies
Our antibody sequencing technology can also accurately complete the sequencing of antibodies labeled with small groups, such as FITC, Biotin, etc., and can also complete the sequencing of antibodies labeled with large protein groups such as GST. Rigorous sequence prediction and stringent sequence verification.
Sample Requirement
- Sample size: > 100 μg monoclonal antibody protein
- Concentration: > 0.5mg/ml
- Purity: The target antibody accounts for more than 80% of the total protein






