The complexity of glycan chains poses a great challenge for accurate qualitative and quantitative glycomic analysis. The study of glycomics relies on the development of glycomic technologies.
In order to accurately mine the characteristic glycoconjugates associated with diseases from multiple glycoconjugates, techniques that can resolve the glycan structure and accurately quantify complex glycoconjugates on glycoproteins in biological systems are needed.
Methods for glycoconjugate analysis in organisms are usually performed by enzymatic or chemical freeing of glycoconjugates from glycoprotein mixtures or individual glycoproteins. Techniques for glycan analysis after freeing are usually lectin microarray techniques, fluorescence capillary electrophoresis, high performance liquid chromatography (HPLC) and mass spectrometry (MS).
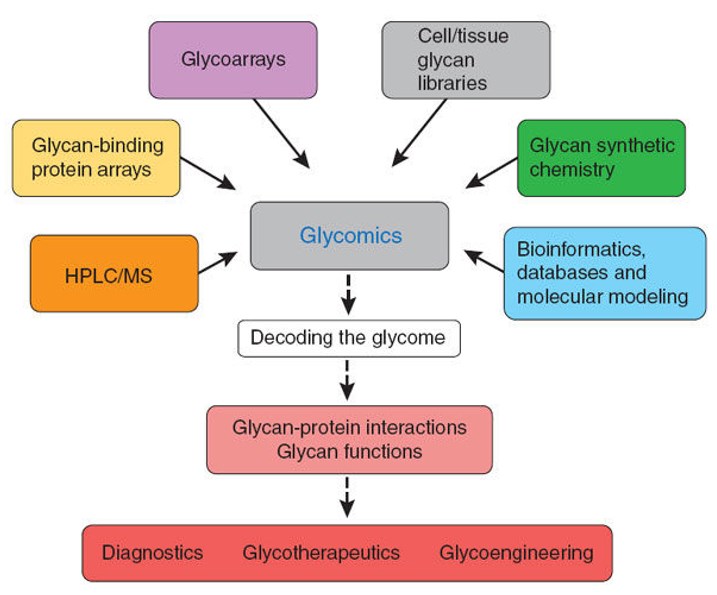
Figure 1. Emerging glycomics technologies
(i) Lectin recognition-based method
This method is fast, simple, sensitive and low cost, and is commonly used for the quantitative analysis of glycosylation of serum and various glycoproteins. However, this type of method relies on known lectin types and care needs to be taken to avoid non-specific binding affecting the accuracy of the results when applied. Other methods are often required when separate quantification of each glycoconjugate composition is required.
(ii) DNA sequencer assisted fluorophore-assisted carbohydrate electrophoresis (DSA-FACE)
DSA-FACE is also a commonly used technique for glycomic analysis, using multiple capillaries to achieve high throughput glycan analysis. However, it is not suitable for the detection of sialic acid-containing glycoconjugates because it requires the removal of sialic acid before detection to reduce the interference of varying amounts of negative charges on the electrophoresis results and to reduce the complexity of the glycomics. Since sialylated glycoconjugates become neutral sugars after the removal of sialic acid and may overlap with the neutral sugars already present in the glycomic group, care should be taken when using this method to consider and verify whether such factors may affect the quantitative results of real glycoconjugate changes in vivo.
(iii) Liquid chromatography (LC)
Liquid chromatography is also a common technique for protein glycosylation analysis. Usually, 2-aminobenzamide (2-AB) is used for derivatization and labeling of the free N-glycan chain. The separation and detection are performed by hydrophilic interaction liquid-ultra performance liquid chromatography (HILIC-UPLC). The method is characterized by high separation, good reproducibility and accurate quantitative results. It can also be used for direct tandem mass spectrometry for glycoform structure identification, and has been widely used for quality control detection and characterization of monoclonal antibody N-glycan profiles. For more complex glycan group analysis such as serum sources, overlapping peaks are easily generated and lead to quantitative errors. Therefore, the UPLC method is currently applied in the development of clinical markers for the analysis and quantification of simple glycomics of individual serum glycoproteins.
(iv) Mass spectrometry
Mass spectrometry has expanded the capability of glycomics research because of its high sensitivity and the ability to provide both structural and quantitative information on glycan chains. The most commonly used biological mass spectra for glycomic analysis are electrospray ionization mass spectrometry and matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS). TOF MS is widely used for the analysis of N-glycan chains because of its rapidity, simplicity, sensitivity, formation of simple spectra dominated by a single charge, and tolerance to a certain level of salt and buffer.
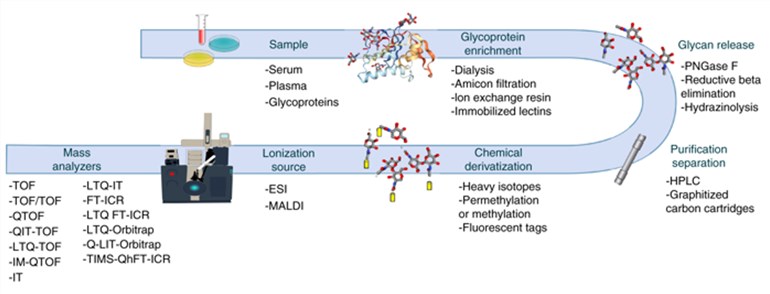
General Overview of a glycomics MS workflow (Rojas et al., 2019)
The instrumental analysis time for MALDI-TOF MS analysis of individual samples is measured in seconds, which enables high-throughput analysis to meet the needs of large samples for clinical sample analysis, and has become a powerful tool for translational studies of glycoconjugate markers to the clinic. MALDI-TOF MS is also widely used in clinical microbiology laboratories for rapid identification of pathogenic microorganisms because of these advantages.
Creative Proteomics use advanced analytical techniques such as mass spectrometry, liquid chromatography, microarray, fluorescence and NMR spectroscopy to identify putative targets. We offer a wild range of glycomics services, including:
- N-Glycan Profiling
- O-Glycan Profiling
- N-Glycosylation Site Occupation Analysis
- O-Glycosylation Site Occupation Analysis
- N-Glycan Linkage Analysis
- O-Glycan Linkage Analysis
- Structural Characterization of Glycans
- Glycopeptides Analysis
- Glycans-related Microarray Assay
- Poysaccharide Analysis
- Peptidoglycan Structure Analysis
Reference
1. Rojas-Macias, M. A., et al. (2019). Towards a standardized bioinformatics infrastructure for N-and O-glycomics. Nature communications, 10(1), 1-10.